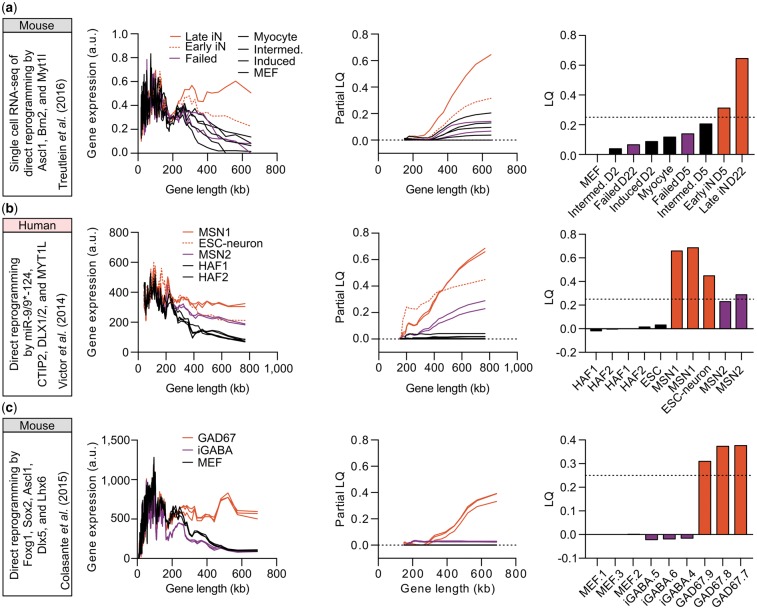
Fig. 3.
LONGO output of somatic cell direct neuronal reprogramming. From left to right, reference for input data, rolling median of gene expression versus length (200 gene bins, 40 gene step), partial LQ and final LQ. (a) Direct reprogramming of mouse embryonic fibroblasts (MEFs) to neurons by transcription factors (Treutlein et al., 2016): MEFs, myocytes, intermediate reprogramming (D2 and D5) and induced MEFs (D2) shown in black; failed reprogramming (D5 and D22) shown in purple; early (D5) and late (D22; dotted-line) successful reprogramming in red. (b) Direct reprogramming of human adult fibroblasts (HAFs) to striatal medium spiny neurons (MSNs) by microRNAs: HAFs from two different cell lines (black); MSNs derived from HAF2 (MSN2; purple); ESC-derived neurons(Gill et al., 2016) (dotted-line) and MSNs derived from HAF1 (MSN1) shown in red. (c) Direct reprogramming of MEFs to iGABA by Foxg1, Sox2, Ascl1, Dlx5 and Lhx6 (Colasante et al., 2015): MEFs (black); iGABA (dark purple); GAD67+ interneurons (red)