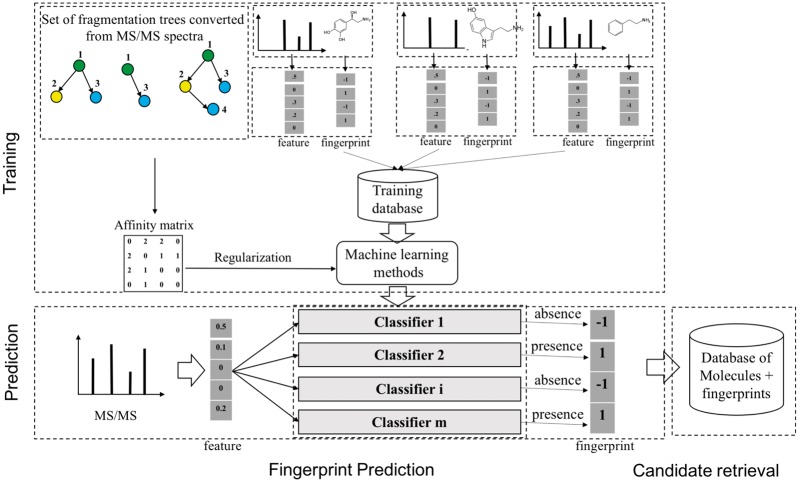
Fig. 2.
A general scheme to identify unknown metabolites based on molecular fingerprint vectors. There are two main stages: 1) fingerprint prediction: learning a mapping from a molecule to the corresponding binary molecular fingerprint vector by classification methods, given a set of MS/MS spectra and fingerprints; 2) candidate retrieval: using the predicted fingerprints to retrieve candidate molecules from the databases of known metabolites. Note that the step of constructing an affinity matrix is optional and is used in L-SIMPLE only