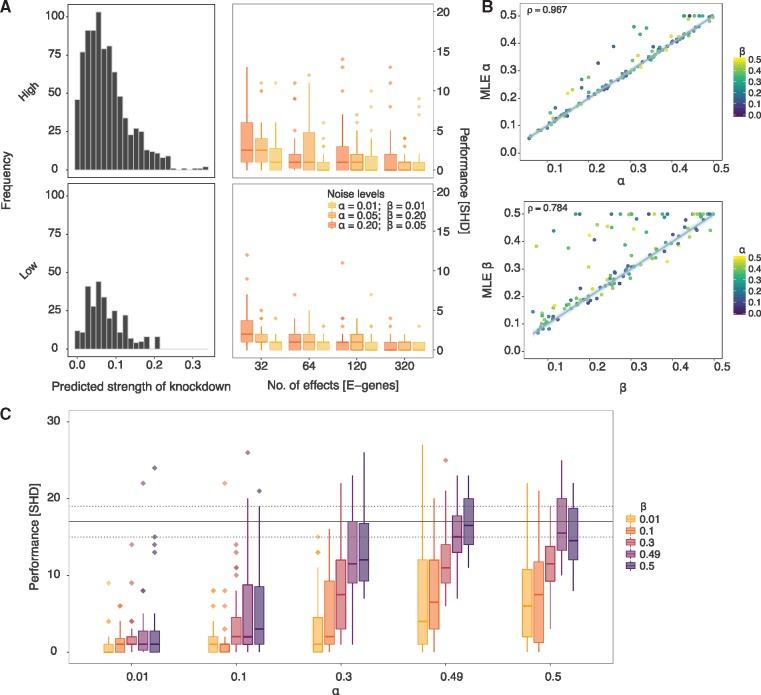
Fig. 4.
Results from simulation study on DAGs. (A) (Left column) Histograms of predicted strength of knockdown depicting the frequency of off-target effects for 30 different DAGs of size N = 8, each sampled from hsa05200 (high density) and hsa00030 (low density) pathways. (Right column) Structural Hamming distance (SHD) measuring the performance (y-axis) of pc-NEMs on simulated data from the corresponding DAGs with varying number of phenotypic effects (x-axis) and noise levels. The box-plots correspond to the inference from data with 32, 64, 120 and 320 effects and known error rates of and (yellow), and (orange), and and (dark-orange). (B) Comparison of 120 different MLE false positive (α) and false negative (β) rates learned using pc-NEMs against true α and β used to generate the data. In estimation of , each point is coloured based on the corresponding value used to generate the data. The error rates are inferred from simulated data with 320 phenotypic effects, without a priori knowledge of the true network structures. (C) Performance (y-axis) of pc-NEMS for network inference without a priori knowledge of error rates, on simulated data with 320 phenotypic effects at different noise levels. The horizontal dashed and solid lines correspond to the lower, median and upper quantile values of SHD for random DAGs