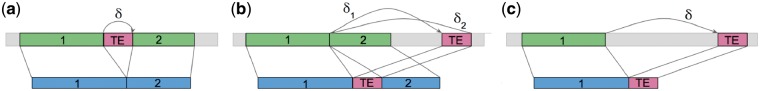
Fig. 2.
Detection of discrepancies caused by TEs. On each subfigure, we plot the reference genome R (top), the contig C (bottom), their matching fragments (blue and green bars for the positions in C and R, respectively) and locations of TEs (violet bars) causing discrepancies in the mapping. The inconsistencies in the alignments are shown by arrows and δ characters. (a) TE is present in R and missing in C. Since δ here is equal to the TE’s length, a specifically chosen breakpoint threshold X transforms classification of this discrepancy from a relocation to a local misassembly (). (b) TE is located inside C but its position in R is significantly away from the rest of C mappings and could also be located on the opposite strand. Original QUAST would treat this situation as two misassembly breakpoints (relocations or inversions) because δ1 and δ2 are usually much higher than X. In contrast, QUAST-LG classifies such pattern as possible TE since it computes , that is again equal to the TE’s length and could be prevailed by appropriate X. (c) TE is the first or the last alignment fragment in C, while its location on R is large distance δ away from the neighboring C fragment. QUAST-LG cannot reliably distinguish this situation from a real relocation/inversion: it would need to be able to recognize TE based on its genomic sequence, which is out of scope of this paper