Figure 1.
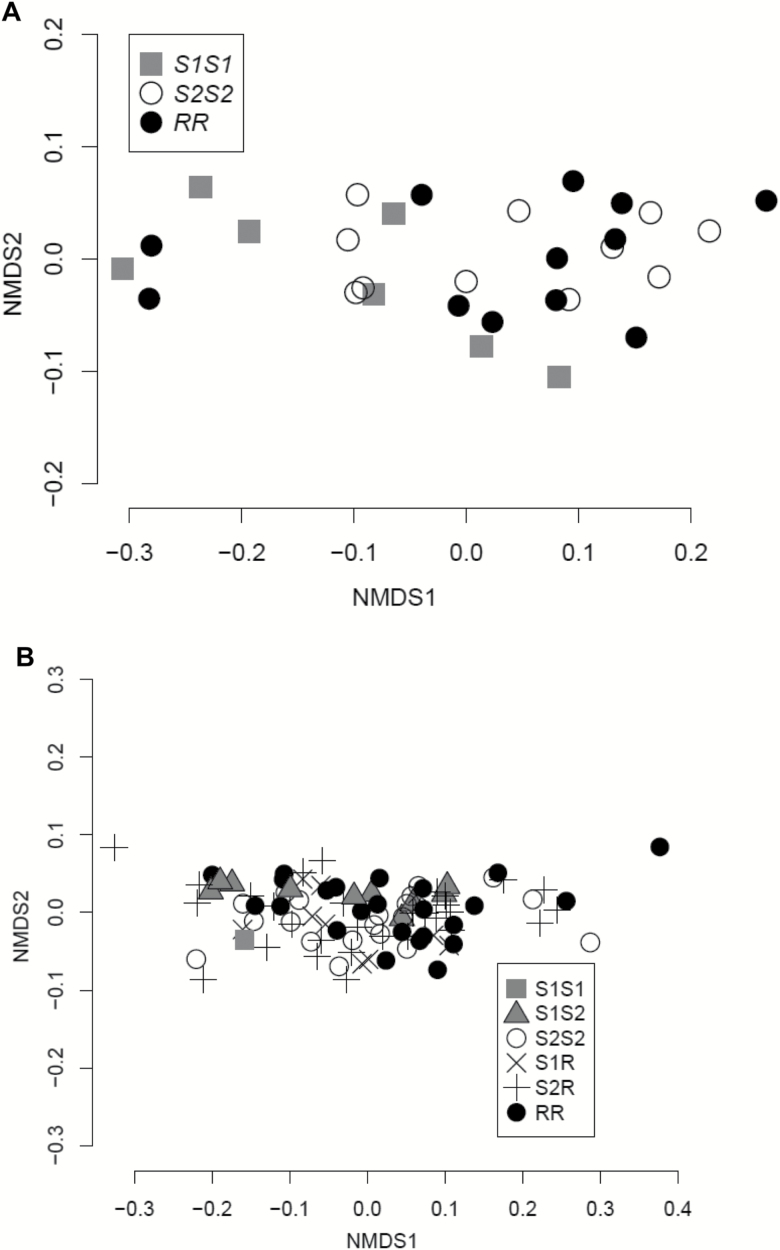
Allelic variation in the GRC region influences the microbiome of BgGUA. (A) OTU composition of the microbiome in homozygous inbred lines (isolated from each other for >3 years; n = 12 RR, 7 S1S1, and 11 S2S2 lines) of BgGUA, as depicted with NMDS of OTUs generated. Overall community composition of S1S1 is significantly different (PERMANOVA Bray–Curtis, P = 0.03) from the other 2 genotypes, although all 3 genotypes show similarly high variability and did not have differing diversity (H). The relative abundance of Micavibrio aeruginosavorus ARL-13 is significantly different in RR lines (uncorrected P = 1.8e-04; α = 0.05/223 = 2.2e-04). (B) OTU composition of the microbiome in outbred BgGUA (2 tanks separated for >1 year) (tank 1: n = 5 S2S2, 7 S1S2, 4 S1R, 14 RR, 19 S2R; tank 2: n = 15 S2S2, 3 S1S2, 6 S1R, 10 RR, 12 S2R), as depicted with NMDS. Allelic variation in the GRC region has no significant influence on the OTU composition or abundance but a significant difference in diversity (H, P = 0.04). All genotypes show similarly high variability.