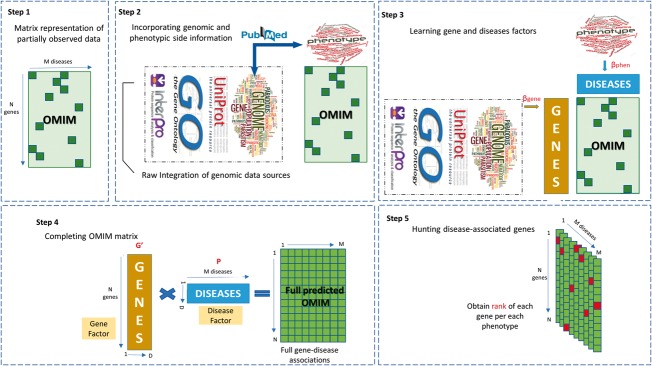
Fig. 2.
Concept of gene prioirtization using matrix factorization. In the first step, a gene-disease asscociation database (OMIM in our case) is represented as a paritally observeed matrix. In the second step, extra information available about genes and phenotypes are prepared to be incorporated into the matrix factorization procedure. Both literature-based phenotypic () and literature-based genomic information are extracted from PubMed. A raw fusion approach is employed to integrate multiple genomic data sources. In the third step, our Bayesian data fusion model jointly learns two thin matrices (Gene and Disease factors) and two link matrix (namely, and ). In fact, this step illustrates the architecture of our matrix factorization approach model(GeneHound) for gene prioritization. In the fourth step, we complete the OMIM matrix using the learned gene and disease factors. Finally, in the fifth step, (GeneHound) ranks all genes in each phenotype column of fully predicted OMIM matrix, separately. For each diseases, genes with the highest predicted value are colored in red