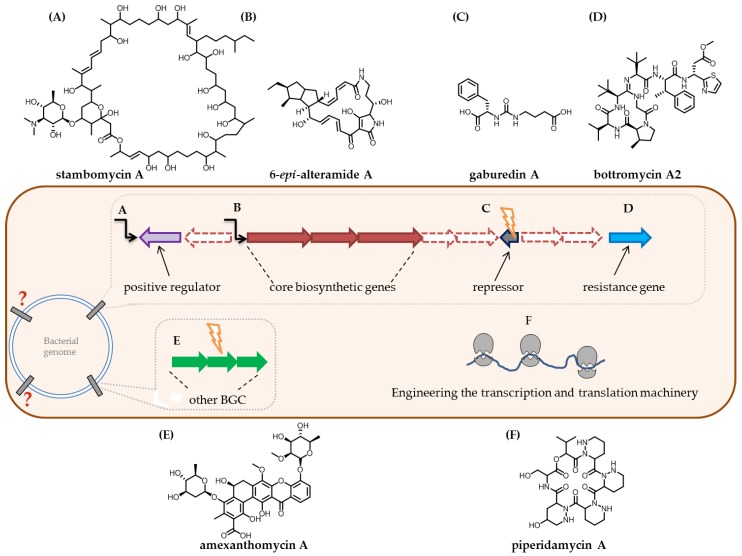
Figure 9.
Generic representation of BGC consisting of core biosynthetic genes, genes for tailoring enzymes (dashed line), positive and negative regulators and self-resistance conferring genes. Strategies can be employed in the native or heterologous host. (A) A promoter is either inserted in front of a positive regulator or an additional copy of the positive regulator with a promoter is inserted in the genome; consequently, a higher concentration of natural product is expected. The macrolide stambomycin A was obtained through constitutive expression of an LAL family regulator [197]. (B) Promoter is inserted in front of the operon of biosynthetic genes enhancing transcription and production of natural product. The polycyclic tetramate macrolactam 6-epi-alteramide A was obtained by introducing the ermE* promoter in front of the hybrid type I PKS-NRPS operon [198]. (C) Repressor gene is disrupted, production of natural product is enhanced. Inactivation of the repressor gbnR in S. venezuelae induced the production of gaburedin A [196]. (D) Resistance gene protects producer strain against its own natural product. During the heterologous expression of bottromycins in S. coelicolor, the replacement of the native promoter by the strong ermE* promoter in front of the efflux pump botT, showed a 20-fold increased production concentration compared to the natively expressed resistance gene [189]. (E) In some cases, knocking out the biosynthetic genes of known compounds in producer strains enhances biosynthesis of other encoded natural products in the genome. Amexanthomycin A was found after knocking out PKS rifA gene responsible for rifampicin biosynthesis from Amycolatopsis mediterranei S699 [190]. (F) Targeted induction of mutation in RNA polymerase and ribosomal proteins by antibiotics can cause upregulation of BGC expression. Mutations in rpsL and rpoB genes activated the silent BGC of piperidamycins from S. mauvecolor, culminating in the isolation of the antibacterial piperidamycin A [209].