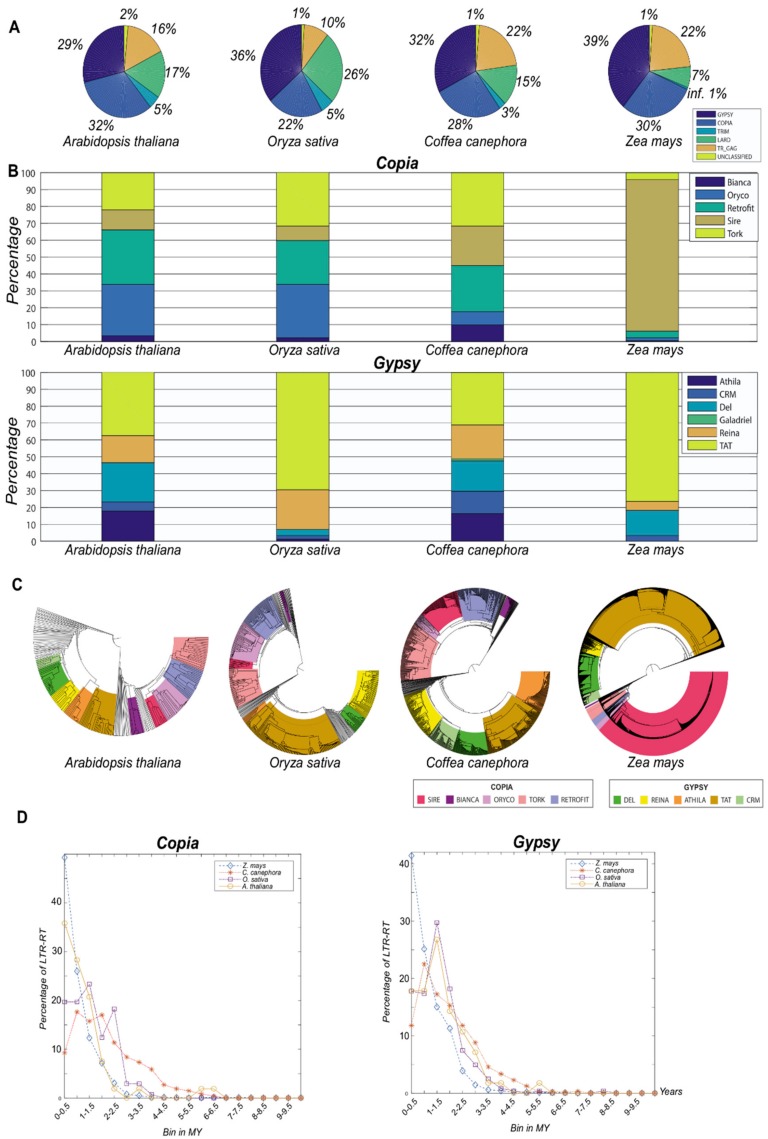
Figure 2.
Inpactor results for the four species tested (Arabidopsis thaliana, Oryza sativa, Coffea canephora and Zea mays) based on LTR_STRUC detection. (A) Initial classification of LTR-RTs into autonomous (Gypsy and Copia) or non-autonomous (Terminal-repeat Retrotransposon In Miniature (TRIM), Large Retrotransposon Derivative (LARD) or Terminal repeat with Gag domain (TR-GAG)); (B) classification of the autonomous elements into lineages showing the variability that can be found in plant genomes; (C) phylogenetic trees using the RT domain; (D) insertion time analysis using autonomous elements (Copia and Gypsy).