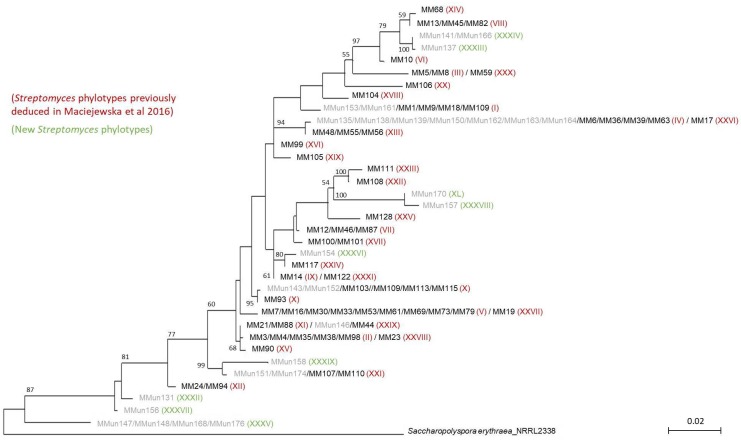
Figure 4.
16S rRNA-based phylogenetic tree of moonmilk-dwelling Streptomyces strains. Strains isolated using the rehydration-centrifugation (RC) method (MMun) are shown in grey, and strains isolated in our previous study (MM) are in black. Phylotype affiliations are indicated in parentheses. The alignment of nearly complete 16S rRNA sequences had 1538 unambiguously aligned positions for 92 strains, but only the 1342 positions without missing nucleotides were used to infer the tree. The evolutionary model was GTR + Γ4 and bootstrap values are based on 100 pseudoreplicates (bootstrap values <50% are not displayed). Saccharopolyspora erythraea was used as outgroup. The scale bar represents 0.02 substitutions per site.