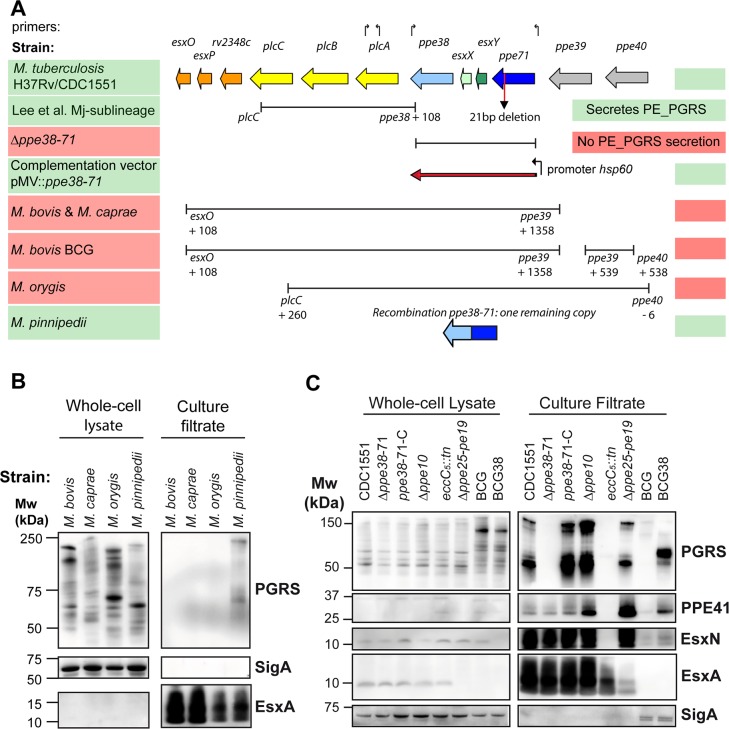
Fig 1. RD5-like genetic deletions in the M. tuberculosis complex and their effect on PE_PGRS secretion.
A) The genetic organization of the RD5 locus in M. tuberculosis strains CDC1551 and H37Rv is depicted in colored arrows. Bars below the genes indicate the size and location of different RD5-like and ppe38-deletions examined in this work. Arrows above the genes indicate primers used in this study to verify the presence of RD5 associated genes, sequences can be found in S4 Table. Functional PE_PGRS secretion is indicated by shading of the strain name in green, while red shading represents strains in which PE_PGRS secretion is not functional (based on immunoblot analysis). Figure adapted from Mc Evoy et al. 2009 with permission [38]. B) Immunoblot secretion analysis of animal-adapted MTBC strains verifies that strains with RD5 deletions do not secrete PE_PGRS proteins. Samples were prepared as described in materials and methods section. C) Immunoblot secretion analysis reveals PE_PGRS secretion defect in BCG, comparable to the M. tuberculosis ppe38-71-deletion strain or a general ESX-5 secretion mutant (eccC5::tn). SigA was used as a loading and lysis control. Some lysis could be found in both BCG and BCG38, but was not markedly different between strains. Please note that these immunoblots correspond to the same pre-cultures as those that were used in the immunogenicity experiment depicted in Fig 5 and therefore also include the Δppe10 and Δppe25-pe19 isolates. Full western blots corresponding to panels depicted in B-C are depicted in S5 Fig and S7 Fig, respectively.