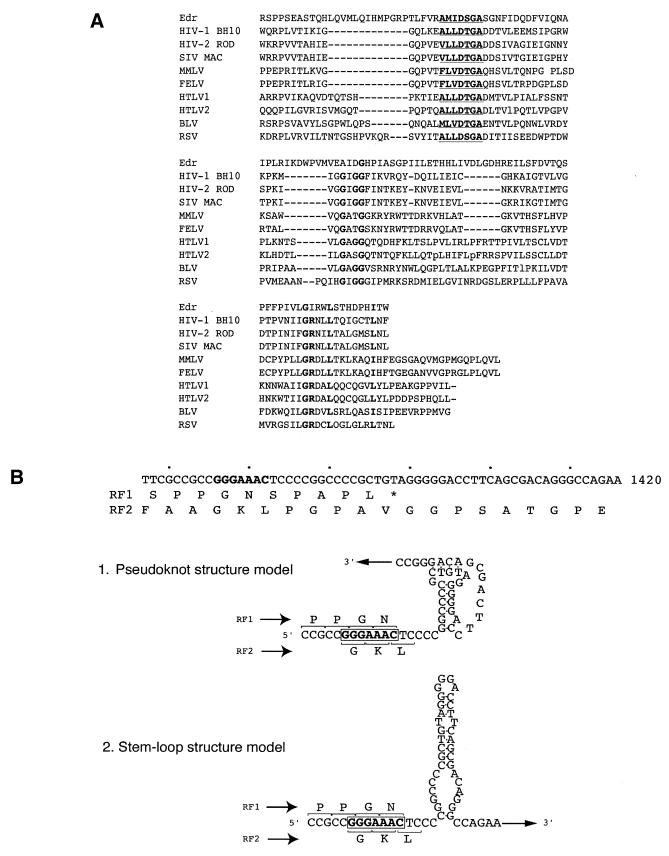
Figure 2.
Identification of retroviral-like motifs in Edr. (A) Amino acid sequence alignment of Edr and retroviral protease motifs. Consensus sequence of aspartyl protease active sites [LIVMFGAC]-[LIVMTADN]-[LIVFSA]-D-[ST]-G-[STAV]-[STAPDENQ]-X-[LIVMFSTNC]-[LIVMFGTA] are underlined. Conserved residues are shown in bold. HIV-1, human immunodeficiency virus type 1 (BH10 isolate); HIV-2, human immunodeficiency virus type 2 (ROD isolate); SIV, simian immunodeficiency virus (MAC isolate); MMLV, Moloney murine leukaemia virus; FELV, feline leukaemia virus; HTLV1, human T-cell leukaemia virus type I; HTLV2, human T-cell leukaemia virus type II; BLV, bovine leukaemia virus. (B) Diagrammatic representation of the predicted –1 ribosomal slippage heptamer and two models of RNA secondary structure downstream from the slippery sequence— a putative pseudoknot and a simple stem–loop structure. The slippage sequence, the potential pseudoknot and a simple stem–loop structures are shown in relation to predicted amino acid sequences of RF1 and RF2. The heptameric –1 slippage sequence is denoted in bold.