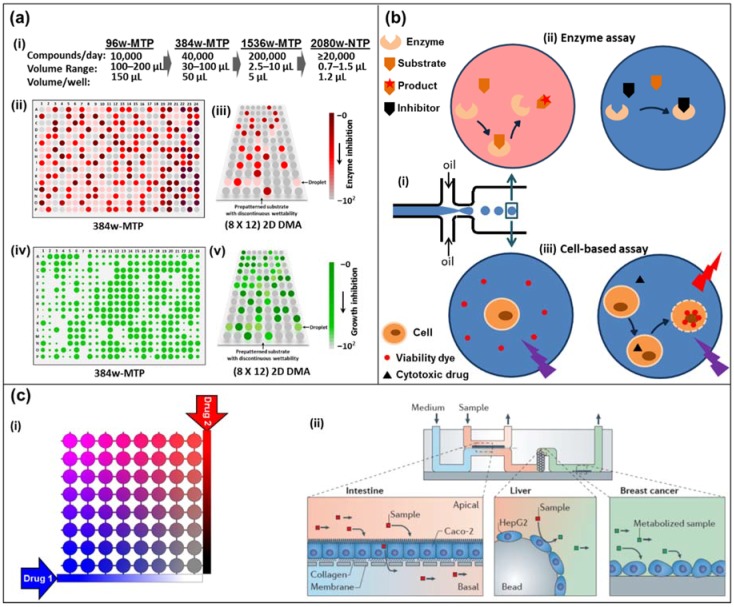
Figure 1.
(a) Miniaturization of high-throughput screening in array formats. (i) Miniaturization of well-plate screening formats. (ii) Schematics of high-throughput screening using a colorimetric assay in 384-well microtiter plate (384w-MTP) and (iii) in 2D droplet microarrays (2D DMAs) on a planar substrate. The intensity of the red-brown color indicates the extent of the reaction (conversely, lack thereof depicts inhibition). (iv) Schematics of high-throughput screening using a cell-proliferation fluorescent marker on 384w-MTP, and (v) in 2D DMA on a planar substrate. The florescence signal is proportional to the extent of cell viability/growth (conversely, no signal means death). (b) Droplet-based microfluidics. (i) Microdroplet formation using the flow-focusing geometry. Droplets are generated at a frequency of 1–1000 droplets/s by shearing an aqueous stream with oil in two directions. (ii) An enzyme and its substrate are encapsulated in a microdroplet (top left). The formation of a colored product can be measured by absorbance. In the presence of an inhibitor in the droplet, product formation is blocked (top right). (iii) A cell and a cell-impermeable viability dye are encapsulated in a microdroplet with (right bottom) or without (left bottom) a cytotoxic drug. Upon cell death, the viability dye binds to the DNA and emits a red fluorescence signal. (c) Diffusion-based microfluidics. (i) Schematic of an 8 × 8 microfluidic diffusion array. Initially, cells (or target molecules) are seeded into the individual chambers (circles). Through a system of reservoirs, pumps and channels, two compounds (e.g., drugs, sensitizers) are diffused into the chambers, each chamber having their individual concentration pair of (Drug 1) and (Drug 2). Over the course of the experiment (e.g., 3–5 days) the chambers can be supplied with fresh media and drug solution while metabolite samples can be collected and analyzed. (ii) A microdevice containing interconnected cell culture microchambers was used to develop a multi-organ model that integrated microfluidic culture of intestinal epithelial cells, hepatocytes and breast cancer cells to simulate absorption, metabolism and activity of anticancer drugs. Adapted by permission from Springer Nature: Nature, Nature Reviews Drug Discovery, Organs-on-chips at the frontiers of drug discovery, [25].