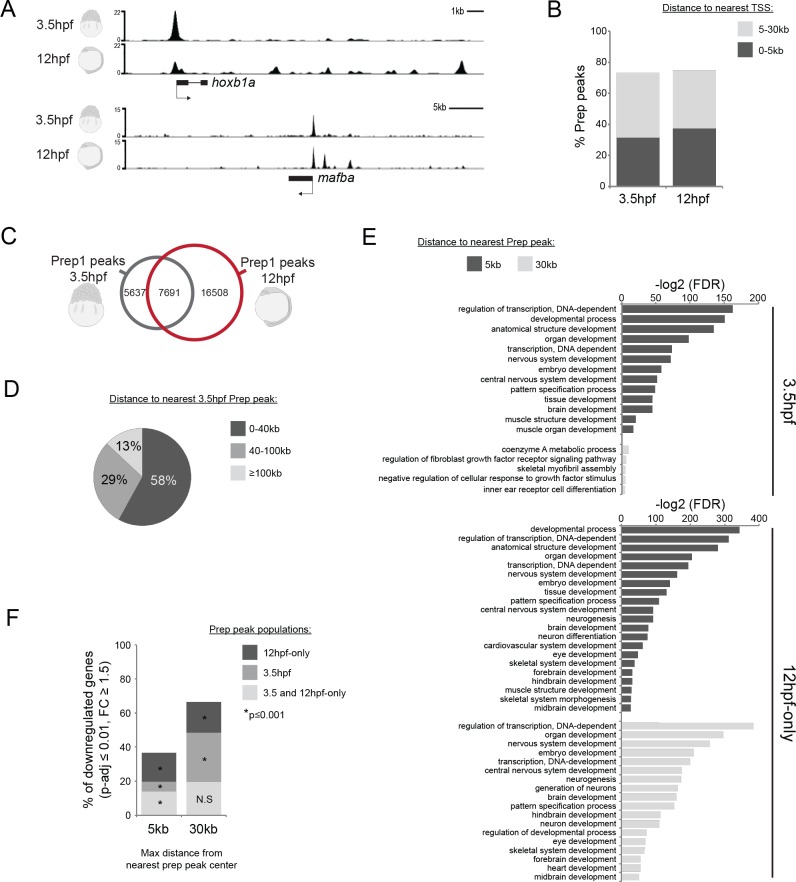
Figure 2. Genomic TALE occupancy is continuously and dynamically associated with the TALE GRN during embryogenesis.
See also Figure 2—figure supplement 1. (A) Representative UCSC browser tracks illustrating Prep binding at the hoxb1a and mafba loci in 3.5 and 12hpf embryos. (B) Graph showing the distribution of Prep3.5hpf and Prep12hpf binding sites relative to TSSs. (C) Venn diagram illustrating co-localization of Prep peaks in 3.5hpf and 12hpf embryos. Two peaks are considered to co-localize if their summits are within 50 bp. (D) Chart illustrating percent of Prep12hpf-only peaks found at various distances from Prep3.5hpf peaks. (E) GO term enrichment for Prep3.5hpf and Prep12hpf-only peaks identified by GREAT using the nearest gene within 5 or 30 kb association rule. In the case of GO terms associated with genes within 30 kb, only select categories are presented, a full list of GO terms is available in Supplementary file 3. FDR = Binomial False Discovery Rate. (F) Graph showing percent of TALE GRN genes (p-adj ≤0.01, fold-change ≥1.5) associated (≤5 or 30 kb) with Prep3.5hpf and Prep12hpf-only peaks. p-values for enrichment above a random set of genes were calculated using the Pearson correlation test.