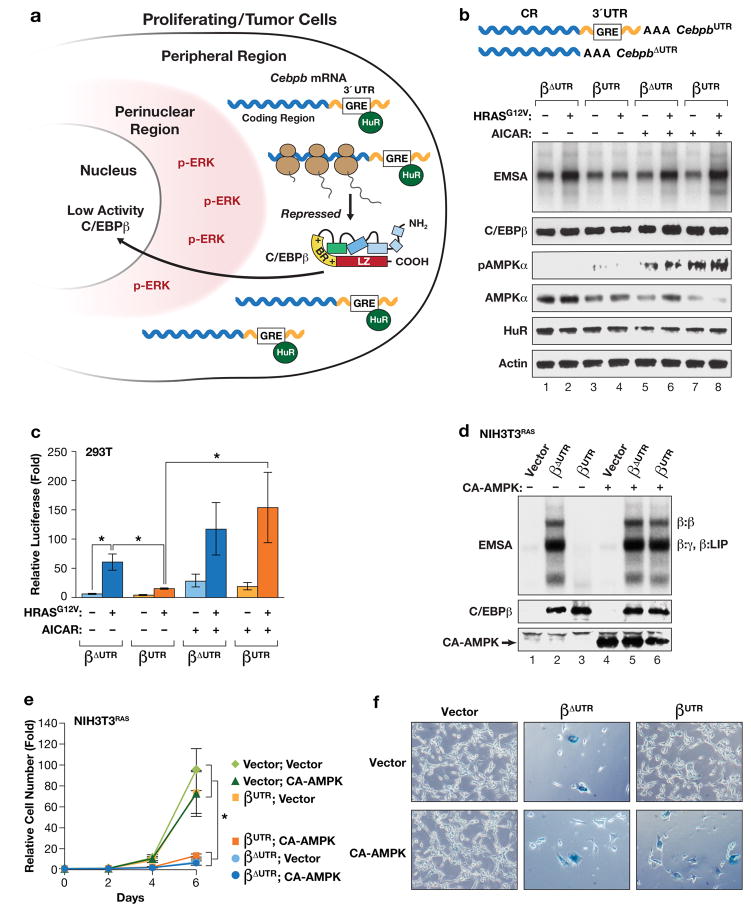
Figure 1.
AMPK signaling abrogates 3′UTR inhibition of RAS-induced C/EBPβ activation. (a) Model depicting “3′UTR regulation of protein activity” (UPA)10 in proliferating and transformed cells. The UPA mechanism involves mutually exclusive localization of Cebpb mRNAs (in the peripheral cytoplasm) and the C/EBPβ kinase, activated ERK1/2 (p-ERK) (in the perinuclear cytoplasm). (b, c) The AMPK agonist AICAR overrides UPA to activate C/EBPβ. The effect of AICAR on C/EBPβ DNA binding (b) and transactivation (c) was analyzed in HEK293 cells. Cells were transfected with C/EBPβ constructs containing or lacking the Cebpb 3′UTR (βUTR and βΔUTR, respectively), without or with HRASG12V, and treated with vehicle or 1 mM AICAR for 16 hr prior to harvest. In (b), nuclear extracts normalized for C/EBPβ levels were analyzed by EMSA using a consensus C/EBP probe. The image was cropped to remove the top and bottom (free probe) portions of the gel. In (c), transactivation assays were performed using a C/EBP reporter, 2XC/EBP-Luc. Luciferase activity, normalized to total protein in each lysate, is plotted as fold increase over the reporter alone. n=3; error bars represent S.E.M. Statistical differences between groups were determined by Student’s two-tailed t test; *p<0.05. (d) Expression of a constitutively active AMPKα1 catalytic subunit (CA-AMPK) reverses UPA inhibition of C/EBPβ DNA binding in RAS-transformed NIH3T3 cells. NIH3T3RAS cells, which express low levels of endogenous C/EBPβ23, were infected with retroviruses expressing βUTR or βΔUTR, without or with CA-AMPK, and assayed for C/EBPβ DNA binding by EMSA. The various C/EBPβ dimeric complexes are indicated. γ: C/EBPγ; LIP is a truncated translational isoform of C/EBPβ61. (e) The same cells were analyzed for proliferation over a 6-day time course. n=3; error bars represent S.E.M. Statistical differences between groups were determined by Student’s t test; *p<0.05. (f) The cells were also stained for the senescence marker, SA-β-Gal. The proportion of SA-β-Gal+ cells in each population is shown in Supplementary Fig. 1c.