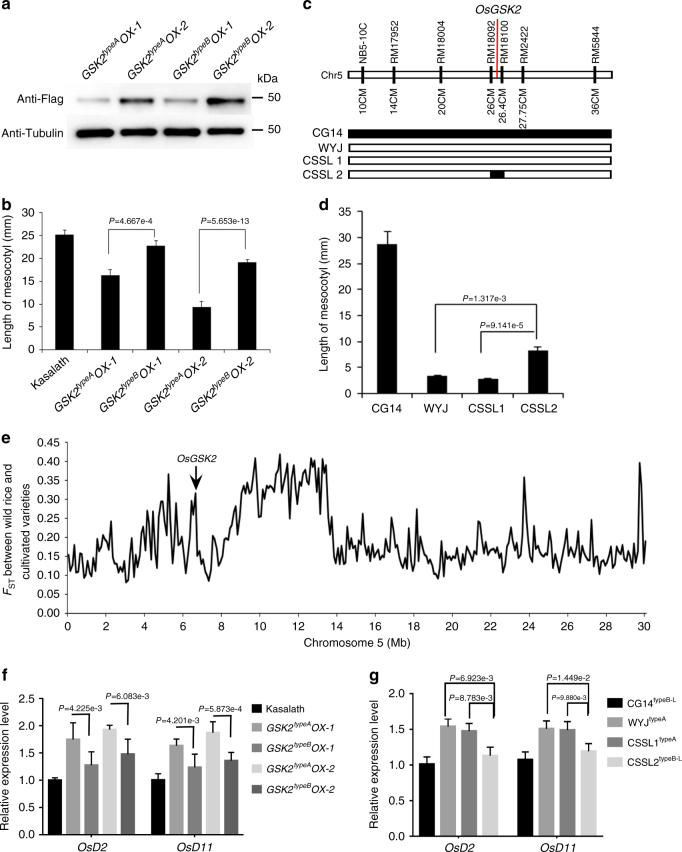
Fig. 2.
The natural alleles of OsGSK2 function in mesocotyl length and BR signaling. a The protein levels of OsGSK2typeA-FLAG and OsGSK2typeB-FLAG in the independent transgenic rice in Kasalath background. b Mesocotyl length of OsGSK2typeA and OsGSK2typeB transgenic rice in a. Kasalath is the background. Error bars are SE (n = 30). P value is determined by Welch’s t test with Bonferroni correction. c Graphical genotypes of the chromosome segment substitution lines used in this study. WYJ is the recurrent parent and CG14 is the donor parent. CSSL1 and CSSL2 are two substitution lines. CSSL1 is a control segregated from the CSSLs population. d The mesocotyl length of the CSSLs and their parents indicated in c. Error bars are SE (n = 25). P value is determined by Welch’s t test with Bonferroni correction. e The level of genetic differentiation (FST) across chromosome 5 between the Oryza rufipogon and cultivated rice. The FST level of OsGSK2 locus is 0.316, and the average FST level in the whole genome is 0.172. f–g The relative transcript levels of OsD2 and OsD11 in OsGSK2 transgenic lines from a and in the CSSLs from c. Total RNAs were extracted from the mesocotyls. The transcript level in WT was defined as “1”. Data are means ± SD (n = 3). P value is determined by Welch’s t test with Bonferroni correction