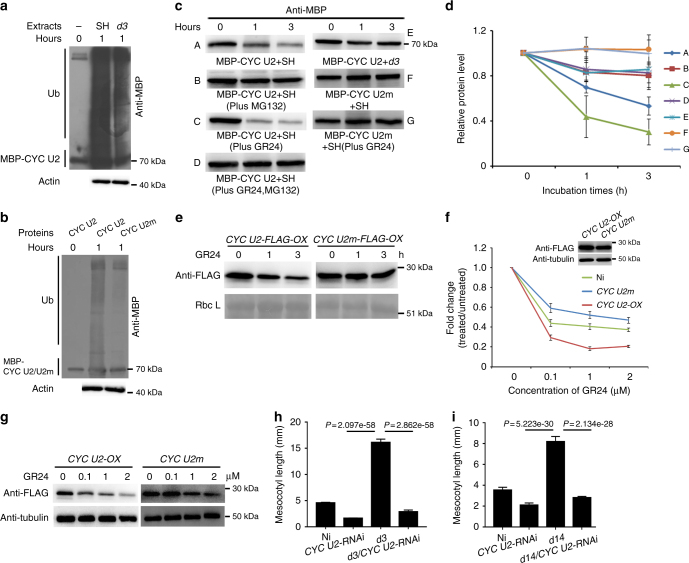
Fig. 5.
SL-inhibited mesocotyl elongation depends on the degradation of the OsGSK2-phosphorylated CYC U2 by D3. a The ubiquitination assay of CYC U2 in SH and d3 mutant. Endogenous actin detected with actin antibody showed the equal amounts of crude extracts from SH and d3 plants incubated. SH is the background of d3. b The ubiquitination assay of CYC U2 and CYC U2m in SH. c The cell-free degradation assays for detecting degradation of CYC U2 and CYC U2m in SH and d3 with or without GR24 treatment. The equal recombinant proteins were incubated with the equal plant crude extracts. d Quantification analysis for c. The relative levels of MBP-CYC U2 and MBP-CYC U2m incubated with the indicated plant extracts at 0 h were defined as “1.” The degradation assay has been independently repeated for three times (means ± SD). e The protein levels of CYC U2-FLAG and CYC U2m-FLAG in the corresponding transgenic rice incubated with 5 µM GR24 at the indicated time points. Rbc L was used as a loading control. f The sensitivity assays of CYC U2 and CYC U2m transgenic rice responding to SL treatments in mesocotyl length. The close-up view indicates the transgenic rice used in (f, g with the similar protein level of the transgenic gene without GR24 treatment. Error bars are SE (n = 15). Ni is the wild-type control. g CYC U2-FLAG or CYC U2m-FLAG protein levels in the mesocotyls of CYC U2-OX or CYC U2m transgenic lines from f. Endogenous tubulin detected with tubulin antibody showed equal loading. h The mesocotyl length of the indicated lines for genetic analysis between CYC U2 and D3. Ni is the wild-type control. Data are mean ± SE. The sample number of Ni, CYC U2-RNAi, d3, and d3/CYC U2-RNAi are 44, 32, 31, and 50, respectively. i The mesocotyl length of the indicated lines for genetic analysis between CYC U2 and D14. Ni is the wild-type control. Data are mean ± SE (n = 35). h, i P values were determined by Welch’s t test with Bonferroni correction