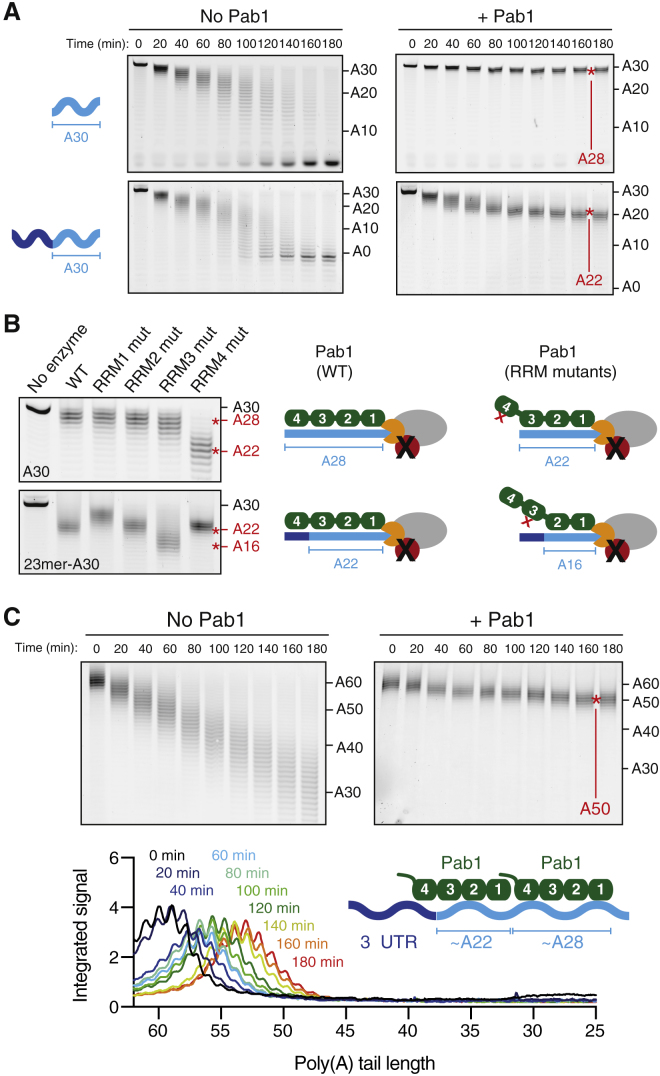
Figure 3.
Pab1 Organization on the Poly(A) Tail
(A) Deadenylation by Ccr4-inactive Ccr4-Not to map Pab1-binding site on A30 and 23-mer-A30 RNA substrates. Red asterisks indicate accumulated product poly(A) tail lengths.
(B) Deadenylation reaction end points (180 min) following addition of Ccr4-inactive Ccr4-Not to A30 (top) and 23-mer-A30 (bottom) RNA substrates in the presence of the indicated Pab1 variants. Red asterisks indicate accumulated product poly(A) tail lengths. Full time courses are shown in Figures S4A and S4B. Models of Pab1 binding to each RNA are shown on the right.
(C) Deadenylation by Ccr4-inactive Ccr4-Not on 20-mer-A60 RNA in the absence or presence of Pab1 (2:1 molar ratio to RNA). Densitometric analysis of the reaction with Pab1 shows that the protected RNA fragment is ∼50–55 adenosines. A model for Pab1-RNA binding is shown.
See also Figure S4.