Figure 5.
Codon Optimality Influences Pab1 Association and mRNA Deadenylation Rate
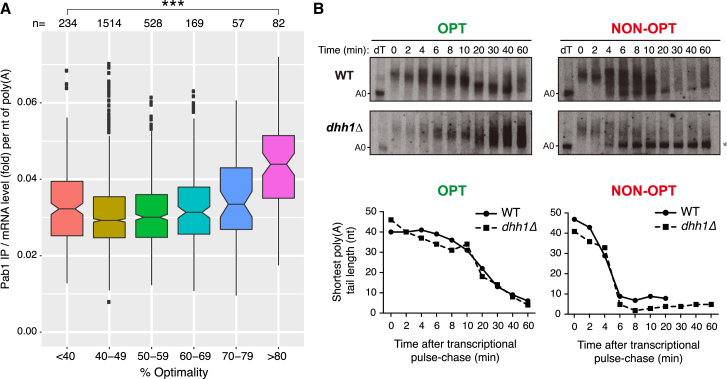
(A) Plot of Pab1-bound mRNA levels relative to total mRNA levels following normalization to poly(A) tail length and binning of mRNAs according to codon optimality. Values were calculated using previously published Pab1 RNA immunoprecipitation sequencing (RIP-seq), total RNA sequencing (RNA-seq), and poly(A) tail length profiling by sequencing (PAL-seq) data. ∗∗∗padj < 10−3.
(B) High-resolution polyacrylamide northern blots and plots of shortest poly(A) tail lengths of the OPT and NON-OPT mRNAs following GAL1 transcriptional pulse-chase experiments in WT or dhh1Δ cells. A0 indicates the migration of a completely deadenylated mRNA species. Asterisk denotes the accumulation of deadenylated mRNA species. The lane labeled dT is the 0 time point treated with oligo dT and RNaseH to indicate the migration position of fully deadenylated mRNA. Representative gels and plots of experiments done in triplicate are shown.