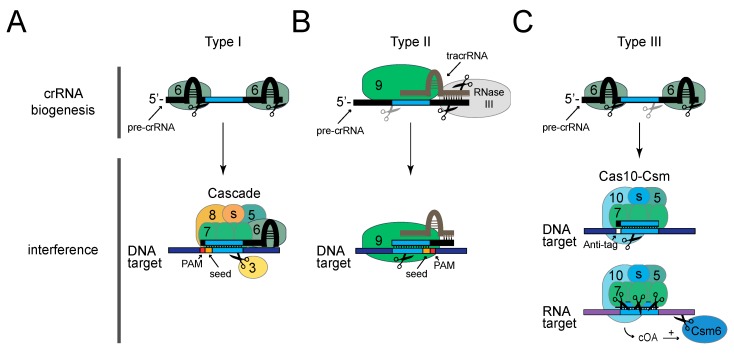
Figure 2.
The three main CRISPR–Cas Types that have been successfully used to edit phages. Shown are protein and nucleic acid requirements for crRNA biogenesis and interference steps in representative CRISPR–Cas systems: Type I-E from E. coli (A), Type II-A from S. pyogenes (B), and Type III-A from S. epidermidis (C). Numbered ovals represent corresponding Cas proteins, and “s” represents one or more copies of a small subunit specific to each subtype. Black scissors represent cleavage points made by the overlapping protein subunit, while grey scissors represent cleavage events catalyzed by non-Cas and/or unknown nucleases. For Type I and II systems, the PAM and seed sequences are represented by red and orange rectangles, respectively. For Type III systems, the crRNA tag is represented by a black square, and the opposing anti-tag is shown as a white square. cOA, cyclic oligoadenylates.