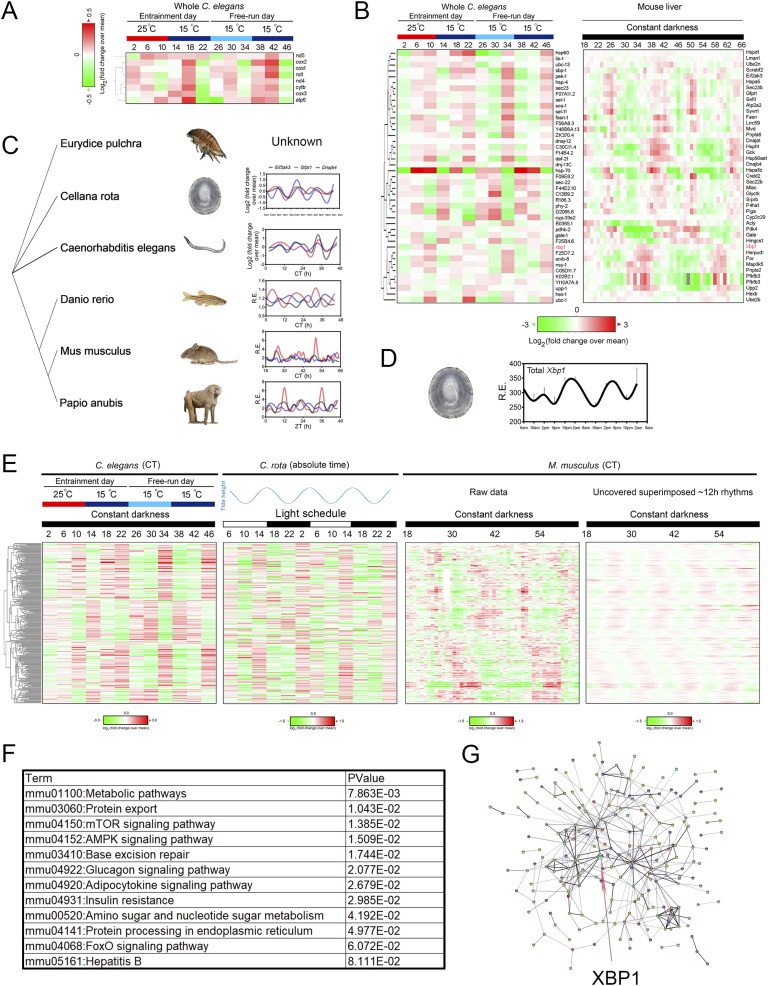
Figure 4.
Twelve-hour rhythms of gene expression are evolutionarily conserved. (A) Heat map of temporal mtDNA-encoded gene expression in temperature-entrained as well as free-running C. elegans compiled from Ref. [75]. (B) Heat map of core ER homeostasis and metabolism related 12-hour cycling gene expression in both temperature-entrained as well as free-running C. elegans (left; compiled from Ref. [75]) and mouse liver under constant darkness (right; compiled from Ref. [8]). Xbp1 is highlighted in red. (C) Phylogenetic tree and relative mRNA expression of Eif2ak3, Gfpt1, and Dnajb4 in C. rota (second row; reported in Ref. [23]), Caenorhabditis elegans (third row; reported in Ref. [75]), Danio rerio (fourth row; reported in Ref. [99]), Mus musculus (fifth row; and reported in Ref. [8]), and Papio anubis (last row; reported in Ref. [100]) during a 48-hour interval. The status of the three genes was not reported for E. pulchra in the study [18]. The data for Papio anubis is double plotted for better visualization. (D) The mRNA level of Xbp1 ortholog in C. rota captured from different times of day from their natural intertidal habitat in the wild as compiled from Ref. [23]. (E) Heat map of all 280 evolutionarily 12-hour cycling gene expression in temperature-entrained as well as free-running C. elegans (left; compiled from Ref. [75]), naturally caught C. rota in tune to 12-hour cycling tidal cues under 12-hour/12-hour natural light condition (middle; compiled from Ref. [23]), and mouse liver under constant darkness (right; compiled from Ref. [8]). (F) Gene ontology analysis of enriched KEGG pathways from these 280 genes. (G) Predicted interactive network construction of these 280 proteins using STRING [102] with XBP1 highlighted by the arrow.