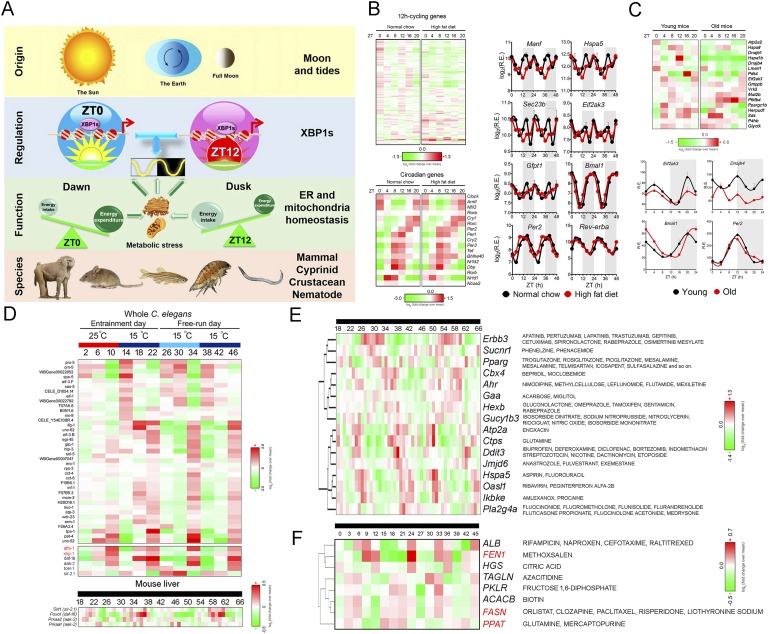
Figure 5.
Mammalian 12-hour clock in diseases and chronotherapy. (A) Diagram summarizing the origin, regulation, function, and species conservation of 12-hour clock. Please see the main text for detailed description of each section. (B) High-fat diet disrupts the hepatic 12-hour cycling, but not circadian gene expression in mice. Heat map (left) and representative log2 normalized expression (right) of key 12-hour cycling and circadian genes under normal chow and high-fat diet conditions are compiled from Ref. [40]. The data are double plotted for better visualization. (C) Disrupted hepatic 12-hour rhythms are associated with aging. Heat map (top) and representative mRNA expression (bottom) of key 12-hour cycling and circadian gene in young and old male mice are compiled from Ref. [152]. (D) Aging-regulating genes are enriched for 12-hour rhythmicity in C. elegans. (Top) Heat map of aging-related 12-hour cycling gene expression in both temperature-entrained as well as free-running C. elegans as compiled from Ref. [75]. Of 62 genes that increase worm lifespan by 10% when knocked down postdevelopmentally [163], 38 (62%) of them showed 12-hour rhythm in temperature-entrained and free-running worms as shown in the heat map. Additionally, positive regulators of aging, including atfs-1, xbp-1, daf-16, aak-2, tcer-1, and sir-2.1, all exhibited 12-hour rhythms of gene expression. (Bottom) Heat map of mammalian ortholog of sir-2.1 (Sirt1), daf-16 (Foxo1), and aak-2 (Prkaa1 and Prkaa2) expression in mouse liver under constant darkness condition as compiled from Ref. [8]. Atfs1 and Xbp1 are highlighted in red. (E and F) Twelve-hour clock–based chronotherapy blueprint. Heat map of temporal mRNA (E) and protein (F) expression of hepatic 12-hour cycling genes with Food and Drug Administration–approved drug interactions as compiled from Refs. [8, 38]. The names of drug are indicated on the right. FEN1, FASN, and PPAT also exhibit 12-hour rhythms of mRNA expression and are highlighted in red.