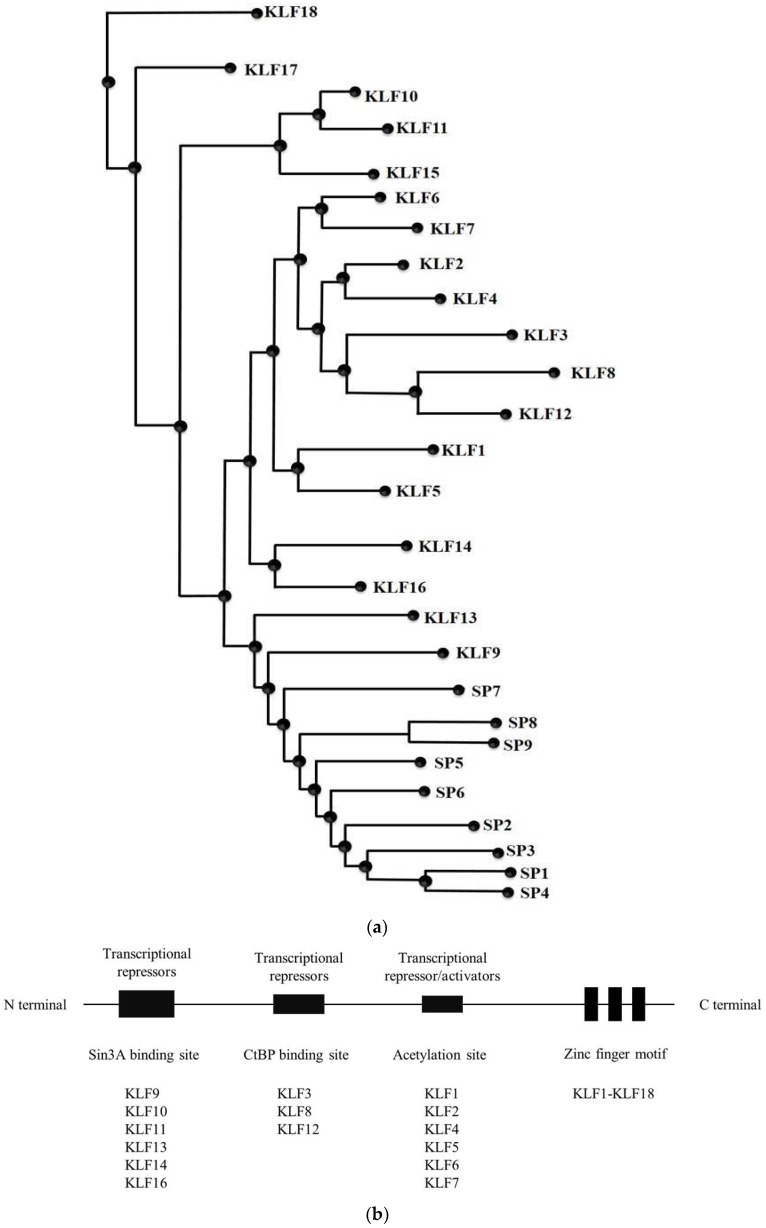
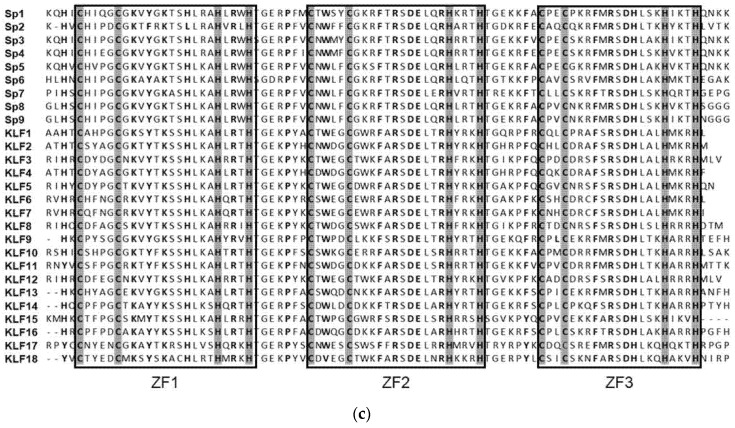
Figure 1.
Phylogenic tree, common structure and zinc finger motif in SP (specificity proteins)/KLF (Krüppel-like Factors) family. (a) Phylogenic analysis based on human protein sequence from NCBI. (b) Structure of KLF family, KLFs isoforms can be divided into 3 groups based on the interaction of their N-terminal site with other proteins for transcriptional co-activators and co-repressors. Group 1 includes KLFs that contain the CtBP binding site. Group 2 includes KLFs that contain the Sin3A interaction domain. Group 3 includes KLFs that interact with acetyl-transferases. KLF15, KLF17, and KLF18 are not included in any of these groups because little is known about their protein interaction motifs and (c) multiple alignments of zinc finger domain of all family members of human Sp/KLF factors. Each protein contains three zinc fingers motifs at the C-terminus, two conserved cysteine residues and two conserved histidine residues for zinc binding are highlighted.