Fig. 1.
OVERVIEW OF THE SRRF ALGORITHM AND OPTIMISING SRRF IMAGING.
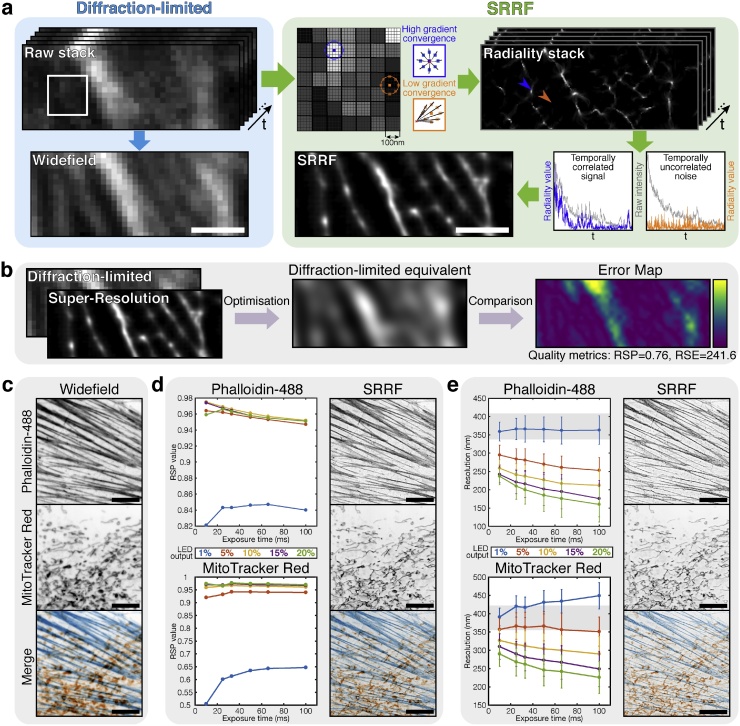
a) Raw data and processing steps in SRRF. The pale cyan box contains the raw, diffraction-limited data series that can be summed to create a single diffraction-limited image. The pale green box contains the SRRF image processing steps. The raw data is split into subpixels (here each pixel from the raw data is split into a 5 × 5 array of subpixels). Examples of the intensity gradients used for the radiality transform are shown for the two highlighted blue and orange subpixels. The blue and orange arrowheads in the radiality stack indicate the location of these subpixels, with their temporal correlations plotted below; coloured plots indicate the radiality variation over time and grey plots indicate the fluorescence intensity variation over time at these locations in the raw data. Scale bar = 1 μm. b) Overview of the SQUIRREL algorithm used for optimising SRRF acquisitions. c) Widefield LED images of Alexa Fluor 488-phalloidin and MitoTracker Red CMXRos (FluoCells Prepared Slide #1, Invitrogen). d) SQUIRREL-calculated RSP (resolution-scaled Pearson’s correlation) values plotted for actin and mitochondria images for different combinations of frame number and exposure time at 5 different LED intensities (indicated as % of maximum output). The highest-quality images (actin: 100 x 10 ms frames at 10% 490 nm LED; mitochondria: 30 x 33 ms frames at 20% 550 nm LED) are displayed alongside. e) SQUIRREL-calculated FRC (Fourier Ring Correlation) mean resolution values (error bars = standard deviation of resolution across the entire image) for the same imaging conditions as in d. Grey shaded regions indicate the mean ± std. resolutions in the widefield images in c. The highest-resolution images (actin: 10 x 100 ms frames at 20% 490 nm LED; mitochondria: 10 x 100 ms at 20% 550 nm LED) are displayed alongside. Scale bars in c-e = 10 μm.