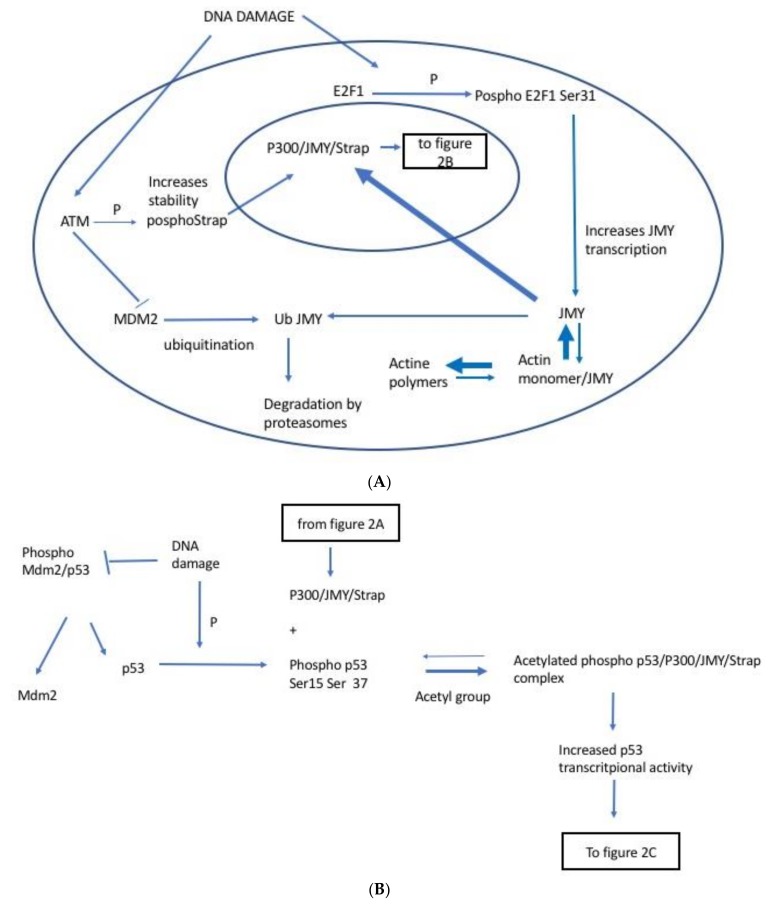
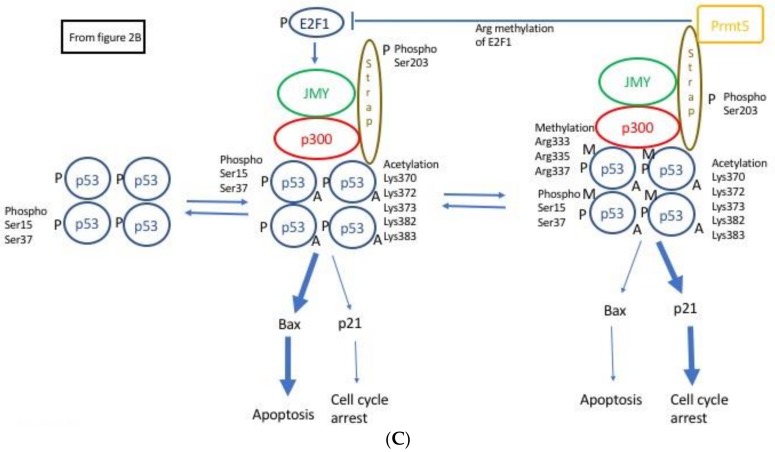
Figure 2.
JMY (Junctional Mediating and Regulating Y protein)/P53 interaction pathway. (A) JMY’s regulation. In unperturbed cells, JMY levels are maintained at a constant state by a balance between transcription and degradation. The latter is controlled by Mdm2 which ubiquitinates JMY leading to its degradation by proteasomes. Following DNA damage, the newly phosphorylated E2F1 induces increased transcription of JMY, while ataxia-telangiectasia mutated (ATM) dampens MDM2 activity. Furthermore, actin monomers form polymers and therefore are no longer available to bind to JMY and sequester it in the cytoplasm. Since it is no longer bound to actin monomers, JMY can then bind to Importin and translocate to the nucleus. This causes the levels of JMY in the nucleus to increase and form a complex with P300. Stability of this complex is further increased by linkage with phosphorylated Stress responsive activator of p53 (Strap). Based on: [18,19,20,21,22]. (B) JMY’s effect on p53. Following DNA damage, p53 is phosphorylated and escapes degradation resulting in upregulation of p53 levels. Phosphorylated p53 binds to the JMY/p300/Strap complex and its transcriptional activity increases. Based on: [9]. (C) Formation of molecular complexes and their effects on apoptosis and cell cycle. Following DNA damage, p53 is phosphorylated and released from MDM2. It binds to the p300/JMY/Strap complex which causes acetylation of five Lysine residues located on the C terminus region of p53. This leads to an increased ability of p53 to transcribe Bax, but not p21, resulting in a preferential activation of the apoptotic pathway over cell cycle arrest. However, when PRMT5 is recruited to Strap that is bound to the JMY/p300 complex, this triggers the methylation of p53 shifting the process away from Bax transcription and apoptosis to increased transcription of p21 and induction of cell cycle arrest. PRMT5 further reinforces this switch by E2F1 inhibition via methylation which reduces JMY’s transcription. Based on and modified from: [9,23,24,25,26,27].