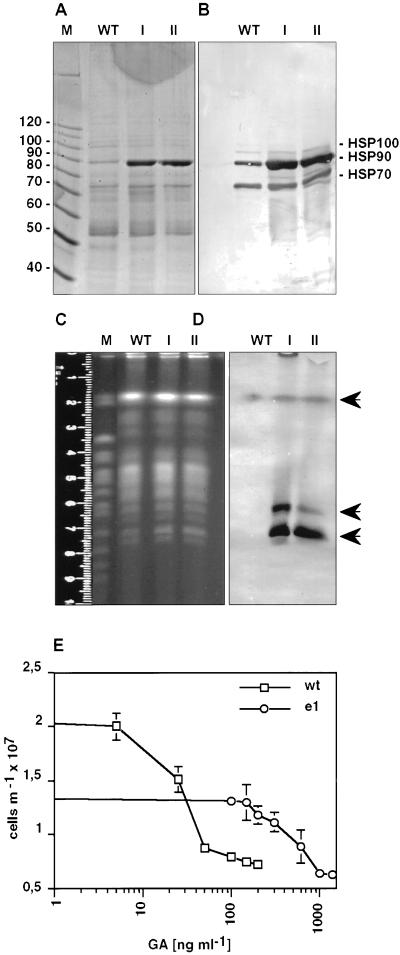
Figure 2.
Amplification of Hsp90 gene copies attributable to geldanamycin selection. (A) SDS-PAGE analysis of wild-type promastigotes (WT) and of two escape strains, I and II, selected at 150 and 200 ng/ml GA, respectively. An equivalent of 106 cells per lane was dissolved in sample buffer and run on a 7.5% gel. The gel was stained with Coomassie blue. The sizes of selected marker proteins (10-kDa ladder; Life Technologies, Gaithersburg, MD) are shown on the left. (B) Immunoblot analysis of the samples described in A. After Western blot, the membrane was probed with a mixture of antibodies directed against Hsp70, Hsp90 and Hsp100. (C) Karyotyping. Chromosomes of L. donovani wild-type and escape strains I and II were separated by pulsed-field gel electrophoresis and stained with ethidium bromide. Saccharomyces cerevisiae chromosomes (strain YPH80) were used as size markers (M). (D) The gel shown in C was subjected to Southern blot, and the membrane was hybridized with a digoxigenin-labeled hsp90 gene probe. The arrows point at bands positive for Hsp90 hybridization. (E) L. donovani wild-type (wt) or escape mutant (e1) cells were seeded at 2 × 106 and incubated for 24 h at increasing GA concentrations. Cell density was measured and plotted against GA concentration.