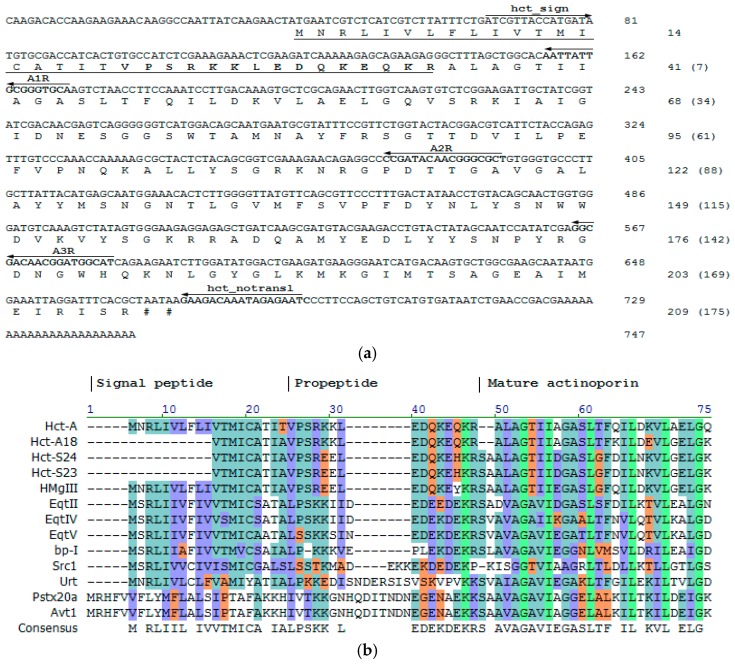
Figure 1.
Structural organization of precursor proteins. (a) Nucleotide sequence of hct-a cDNA accompanied by deduced amino acid sequence. The signal peptide sequence is underlined, the propeptide sequence is bolded and underlined. The protein sequence is numbered starting from the presumed initiation methionine residue. The numbering of the mature polypeptide is in parentheses. The nucleotide sequences corresponding to primers A1R, A2R, A3R, hct_sign, and hct_notransl are bolded and are indicated by arrows. (b) Multiple sequence alignment for four actinoporin precursors from H. crispa and other sea anemones: HMgIII (Q9U6X1) from H. magnifica; EqtII (P61914), EqtIV (Q9Y1U9), and EqtV (Q93109) from A. equina; bp-1 (C5NSL2) from A. asiatica; Scr1 (Q86FQ0) from S. rosea; UcI (C9EIC7) from U. crassicornis; PsTX-20A (Q8IAE2) from P. semoni; Avt1 (Q5R231) from A. villosa; non-similar amino acids are shown on white background, weakly similar—on light-grown, conservative—on light-blue, block of similar—on purple, identical—on light-green colors. Gaps are represented by dashed lines. The boundaries of the signal peptide, prepropeptide, and N-terminal fragment of the mature protein are shown by vertical lines.