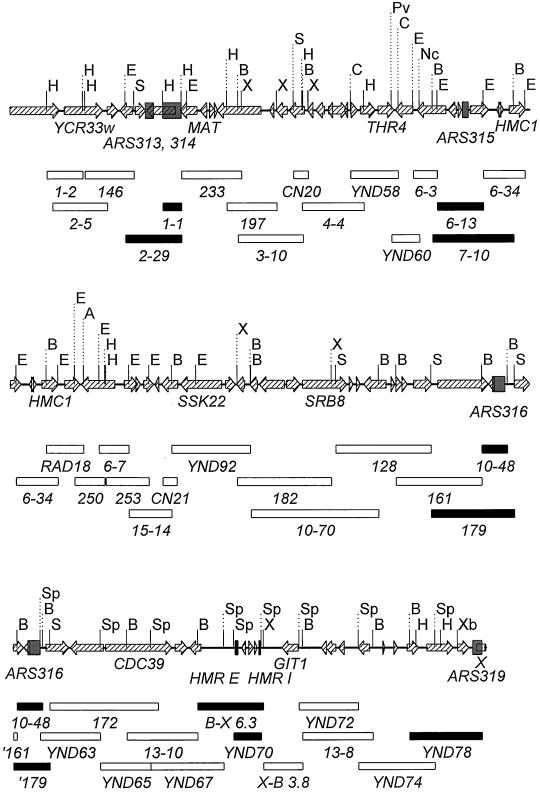
Figure 1.
Map of the rightmost 136 kb of chromosome III. Three overlapping segments of this region are shown with open reading frames indicated by arrows on the map lines and restriction sites used for subcloning indicated above the map lines. Open reading frames are based on the September 2000 release of the chromosome III sequence, and names indicated are from the Saccharomyces Genome Database (February 2001). Only restriction sites that mark the boundaries of subclones are shown; additional sites exist for each of these enzymes. The top segment extends from nucleotide 181,264-231,267, the middle segment from 226,264-276,191, and the bottom segment from 271,271 to the right end of the chromosome at nucleotide 316,613. The positions of ARS elements, identified by the subclones shown in this figure and further refined by subcloning shown in Figure 2, are indicated by shaded boxes. The open box at the right end of the lower segment indicates the position of the subtelomeric X, A, B, C, and D sequences (Louis et al., 1994). These subtelomeric sequences are followed by 93 bp of the G1–3T telomeric repeat. Boxes below the map represent subclones prepared in the vector pRS306 and analyzed for ARS activity, with open boxes representing Ars− subclones and filled boxes representing Ars+ subclones. With the exception of subclones YND58, YND60, RAD18, YND78, and YND79, described in Table 1, all subclones were derived from chromosome III inserts in bacteriophage λ or cosmid vectors prepared from strain AB972 by Riles and Olson (1993) and used in the chromosome III sequencing project (Oliver et al., 1992). Restriction enzyme abbreviations: A, AatII; B, BamHI; Bn, BstNI; Bs, Bst1107; Bw, BsiWI; C, ClaI; E, EcoRI; H, HindIII; K, KpnI; Pv, PvuII; Nc, NcoI; Rv, EcoRV; S, SalI; Sc, SacI; Sh, SphI; Sp, SpeI; Ss, SspI; X, XhoI; and Xb, XbaI.