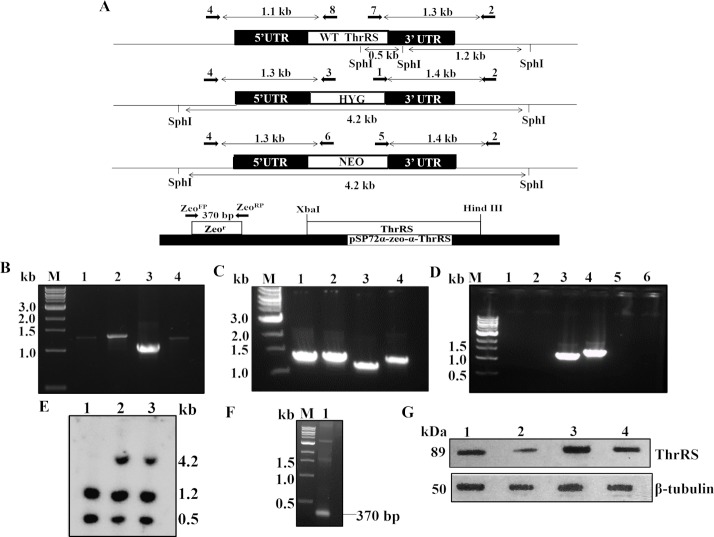
Fig 3. Generation of heterozygous mutants of LdThrRS.
(A) Map of LdThrRS genomic locus and pSP72α-zeo-α-ThrRS episomal construct is shown with the position of the primers used for confirmation of WT and mutant parasites by PCR-based analysis along with the expected band sizes. Primer pairs are shown in Table 2. Primer 4 was designed as a forward primer to match the upstream region of LdThrRS gene, and primers 8, 3 and 6 were designed internal to LdThrRS, HYG and NEO coding regions, respectively. Primer 2 was designed as a reverse primer to match the downstream region of LdThrRS gene and primers 7, 1 and 5 were designed as forward primers, internal to LdThrRS, HYG and NEO coding regions, respectively. Forward primer ZeoFP and reverse primer ZeoRP were designed to match the upstream and downstream region of zeocin resistance gene. Genomic DNA from WT, heterozygous parasites (ThrRS/NEO or ThrRS/HYG) was used as a template for PCR analysis. (B) The specific fusion of the replacement cassette(s) was checked with NEO and gene-specific primers as reported in Table 2 and Fig 3A. Lane 1 (Primers 4 and 6); Lane 2 (Primers 5 and 2); Lane 3 (Primers 4 and 8) and Lane 4 (Primers 7 and 2). (C) The specific fusion of the replacement cassette(s) was checked with HYG and gene-specific primers. Lane 1 (Primers 4 and 3); Lane 2 (Primers 1 and 2); Lane 3 (Primers 4 and 8) and Lane 4 (Primers 7 and 2). (D) WT genomic DNA was used as a negative control. The bands corresponding to the WT gene were obtained in Lane 3 and 4. No bands were observed after using NEO specific primers, Lane 1 (Primers 4 and 6); Lane 2 (Primers 5 and 2) and HYG specific primers, Lane 5 (Primers 4 and 3); Lane 6 (Primers 1 and 2). M indicates the molecular size marker in kb. (E) Southern blot analysis of genomic DNA from wild-type (WT) (Lane 1), ThrRS/NEO (Lane 2) and ThrRS/HYG (Lane 3) parasites. Genomic DNA from WT, ThrRS/NEO and ThrRS/HYG parasites were digested with SphI, separated on a 0.6% agarose gel and probed with 3’UTR of LdThrRS gene. (F) PCR analysis of rescue mutants (ThrRS/NEO[pThrRS+]). The specificity of recombination was checked with Zeor specific primers, Lane 1 (Primers ZeoFP and ZeoRP). M indicates the DNA molecular size marker. (G) Western blot analysis of WT (Lane 1), heterozygous parasites (ThrRS/NEO) (Lane 2), ThrRS overexpressors (WT[pThrRS+]) (Lane 3) and rescue mutant (ThrRS/NEO[pThrRS+]) (Lane 4) parasites. β- tubulin was used as a loading control.