Figure 3. NFRe promotes nodule organogenesis in Lotus japonicus.
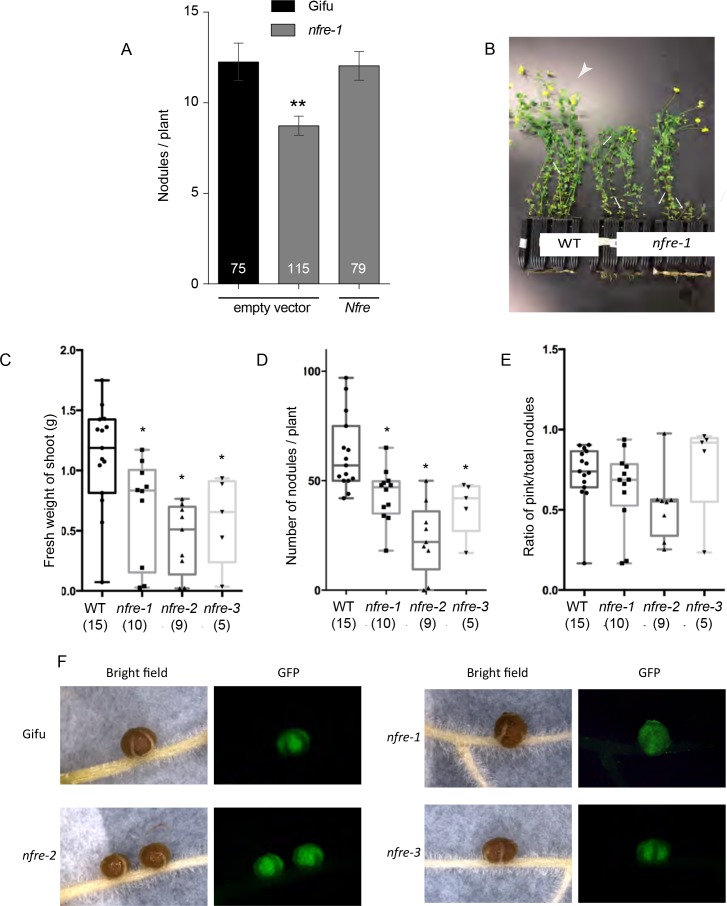
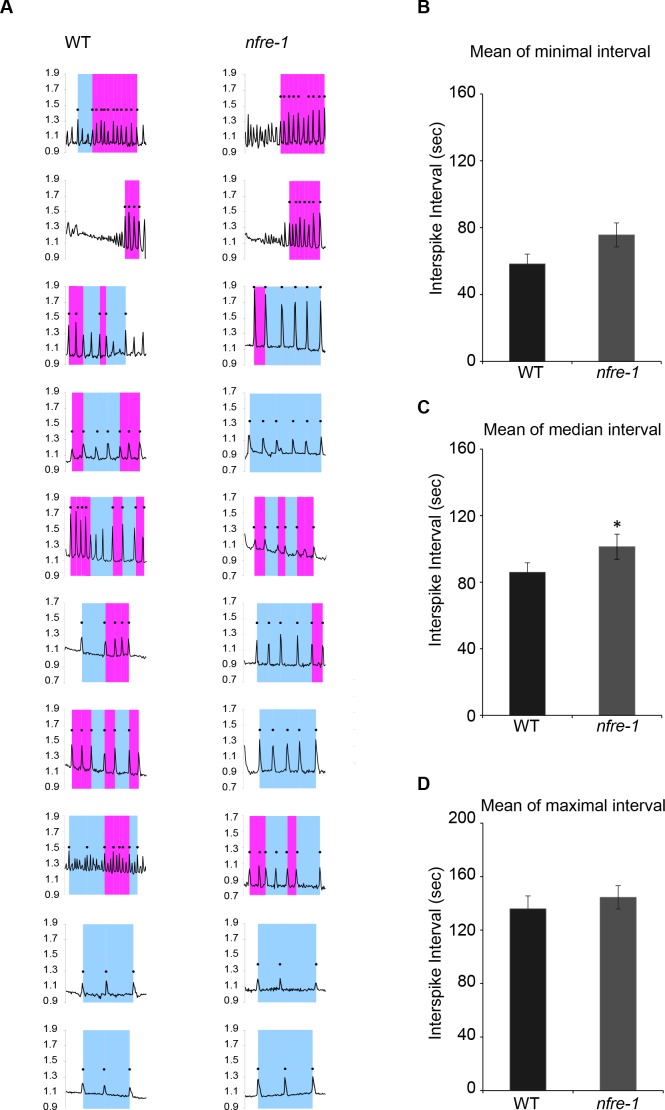
(A) Greenhouse-grown nfre plants formed fewer root nodules compared to WT when analysed at eight wpi with M. loti. (B) Agar plate-grown nfre plants form fewer primordia than WT at 5 wpi with M. loti. (C) The nfre mutants and wild type plants form similar number of short and long root hair infection threads at 9 and 14 dpi. (D) Representative nuclear calcium oscillations (spiking) induced by R7A Nod factor (10–8 M) in wild type and nfre mutant root hairs. Ca2+ oscillations are presented as ratiometric values between YFP and CFP signals detected on the basis of the NLS-YC3.6 Ca2+sensor. (E) The inter-spike interval of nfre-1 mutant is significantly longer than that of WT. (F) Venn diagrams of Nod factor up- and down-regulated (parentheses) genes detected in the susceptible zone at 24 hr after treatment. The values given at the bottom of columns in (A) and (C) represents the number of plants analysed. Error bars represent standard error of the mean. *p<0.05 and **p<0.01, Student´s t-test compared to wild type.
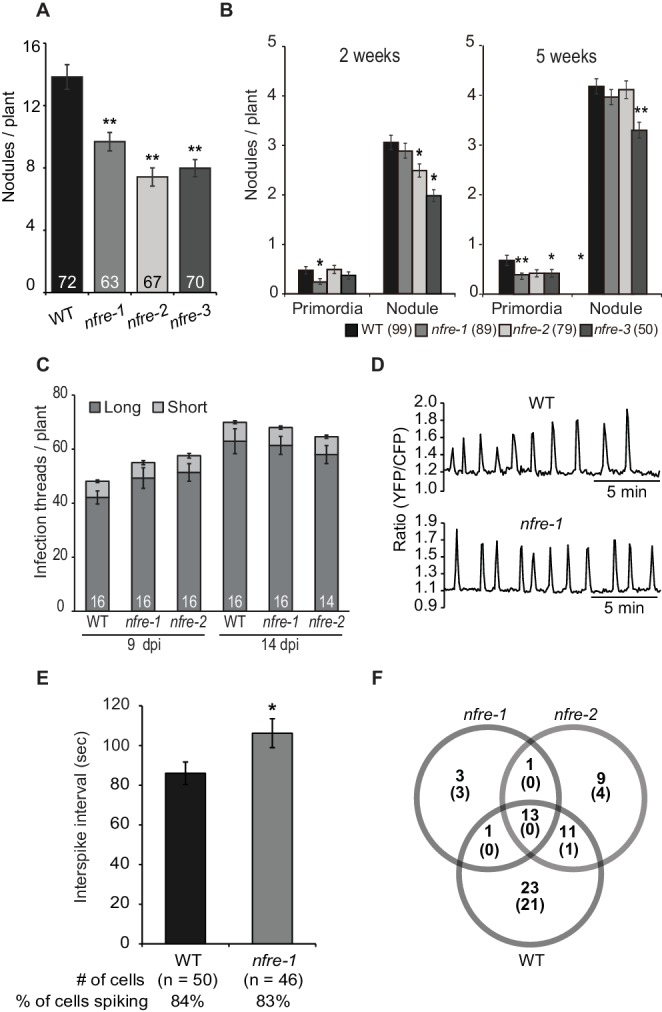
Figure 3—figure supplement 1. The phenotype of wild-type and nfre plants.
Figure 3—figure supplement 2. Pattern of nuclear calcium oscillations in wild-type and nfre-1 mutant.
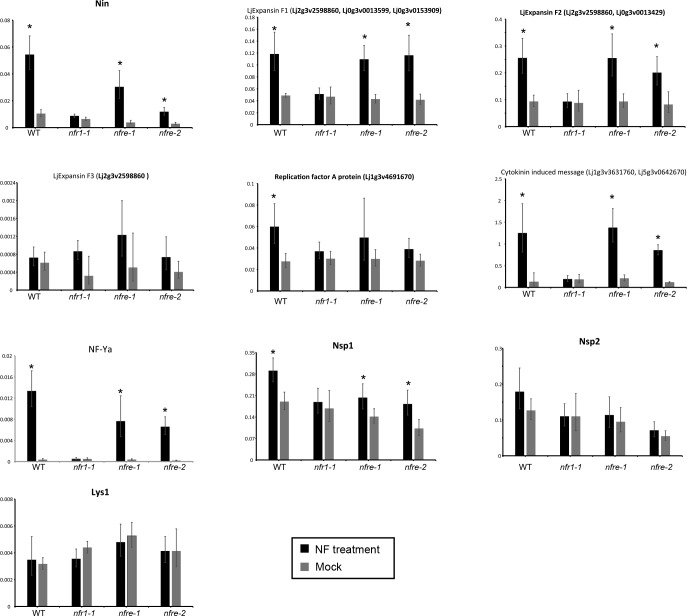
Figure 3—figure supplement 3. Transcript levels of selected genes measured by quantitative RT-PCR in Mock, or Nod factor-treated wild type, nfr1-1 and nfre mutant roots.
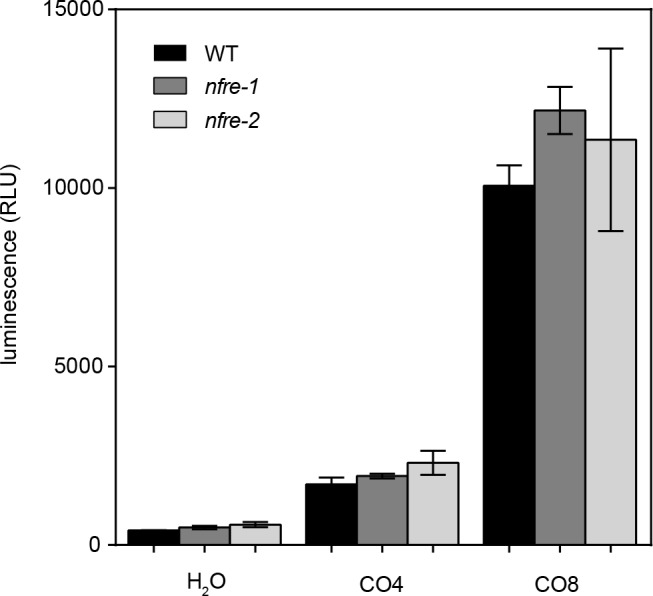
Figure 3—figure supplement 4. Chitin oligomers elicit production of similar ROS levels in nfre and wild type roots.