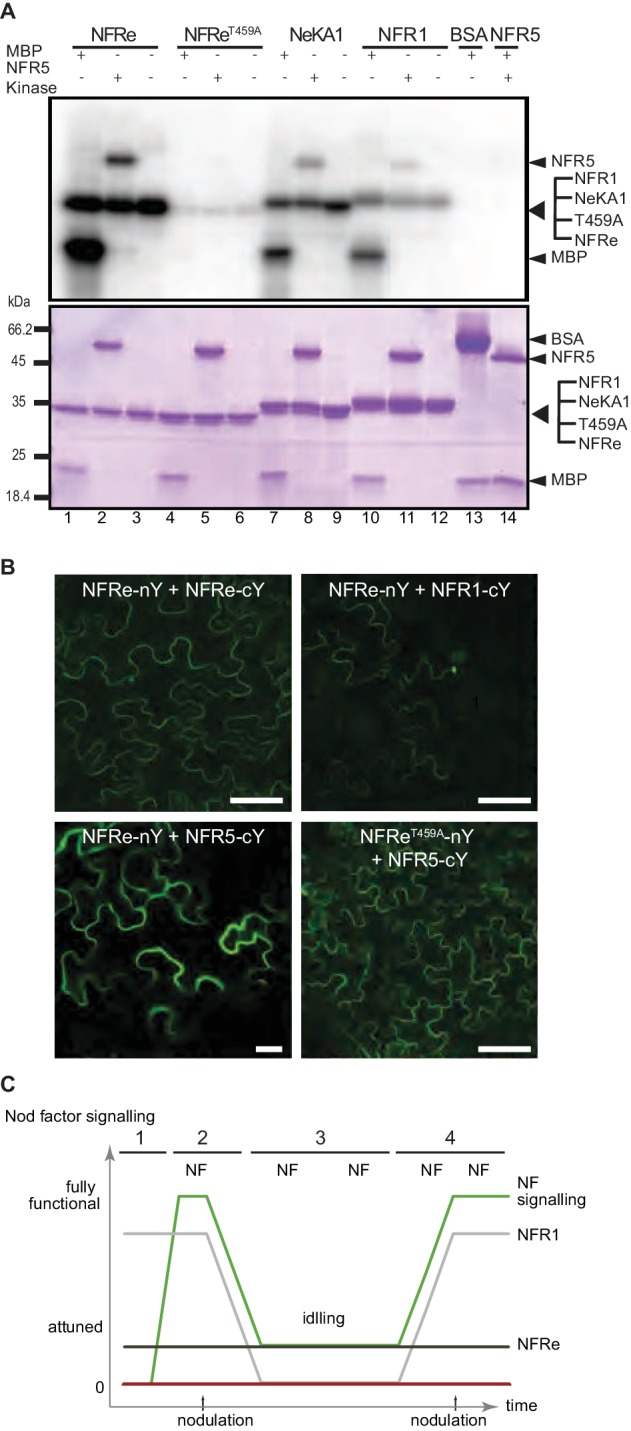
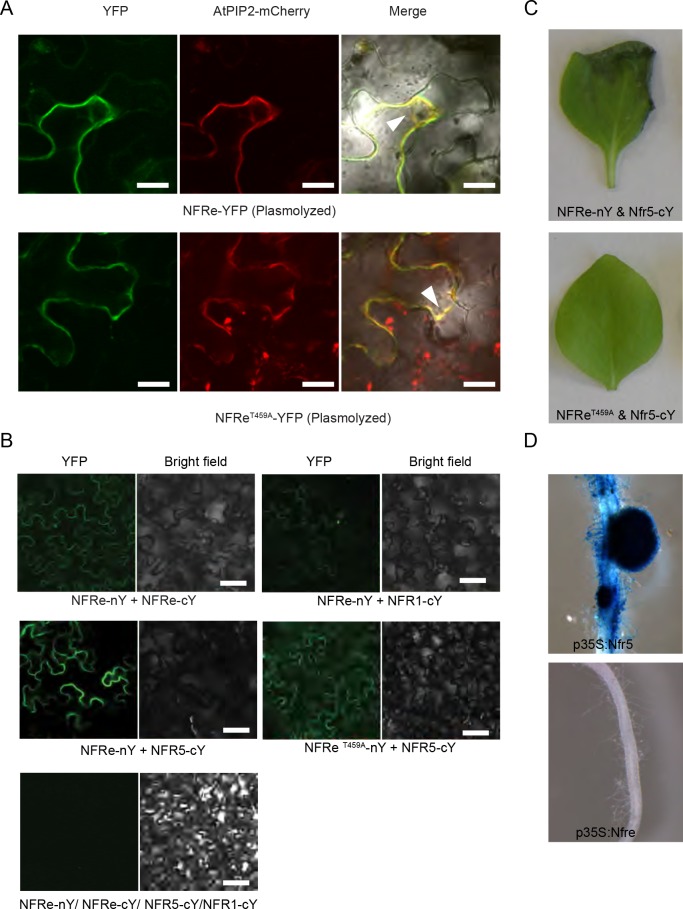
Figure 4. NFRe signalling is dependent of NFR5.
(A) The NFRe kinase phosphorylates NFR5 kinase, whereas the NFReT459A shows no phosphorylation activity. The kinase of NeKA1 receptor in which the activation segment of NFRe was swapped with the corresponding region of NFR1 also phosphorylates NFR5 kinase. NFR1 kinase serves as positive control for NFR5 kinase transphosphorylation. Bovine serum albumin and NFR5 kinase domain are negative controls. (B) Bimolecular fluorescence complementation (BiFC) of YFP signal indicates protein-protein interactions in tobacco leaves. NFRe forms homomers (NFRe-nY +NFRe cY), and heteromers with NFR1 (NFRe-nY +NFR1 cY), or NFR5 (NFRe-cY +NFR5 nY). Formation of heteromeric complexes with NFR5 is not dependent on an active NFRe kinase (NFRe T459A -nY + NFR5 cY). (C) Working model of Nod factor signalling (green line) in the susceptible zone ensuring an efficient nodulation on the expanding root system. NFRe (dark grey line) has a constant expression in the epidermal cells of the susceptible zone. NFR1 (light grey line) dominates the uninoculated root in terms of expression level and spatial distribution (1). Once the symbiotic process is initiated by the Nod factor (NF), the expression of NFR1 is rapidly downscaled (2). A sustained expression of NFRe in the epidermal cells ensures an idling signalling in the susceptible zone, keeping the expanding root system tuned in to rhizobia (3). NFR1 acts as a master switch triggering recurrent symbiotic events in a fast and efficient manner from NFRe-attuned epidermal cells (4).