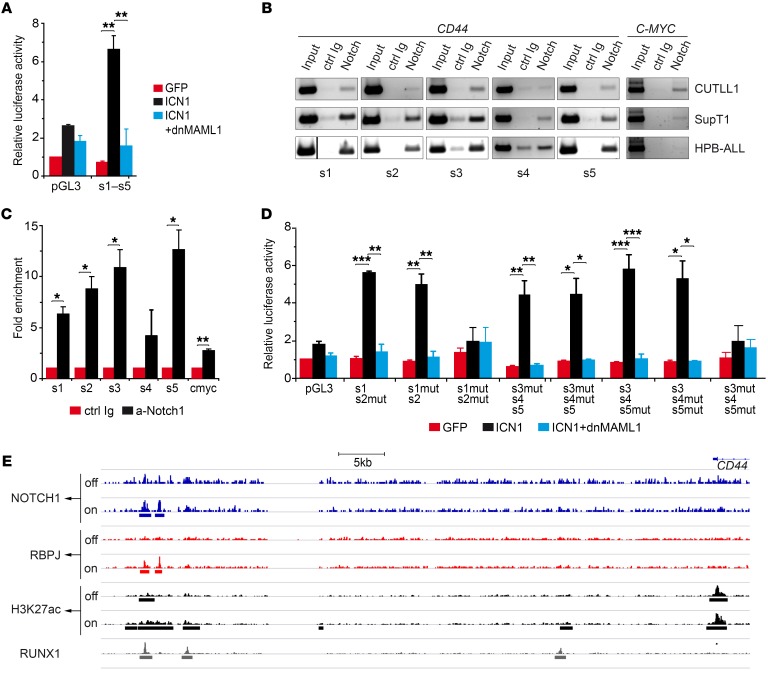
Figure 4. CD44 is a CSL/MAML-dependent direct target of NOTCH1.
(A) Luciferase reporter assays of SupT1 T-ALL cells cotransfected with a reporter construct containing a genomic CD44 region (–4.8 to –3.5 kb) spanning 5 putative CSL-binding sites along with ICN1-encoding retrovirus plus or minus retrovirus encoding dnMAML1 or with GFP-only control. Data are represented as mean fold induction ± SEM over luciferase activity of control cells cotransfected with an empty reporter vector (pGL3) and the GFP-only vector (n = 3). s1–s5, CSL-binding sites 1–5. (B) Representative ChIP assay performed either with anti-NOTCH1 Abs (anti-cleaved NOTCH1, for CUTLL1 and HPB-ALL, or anti–C-terminal NOTCH1, for SupT1) or with the corresponding control Ab. PCR amplification was done on input and immunoprecipitated DNA with primer pairs spanning each of the 5 CD44 CSL sites or the reported c-MYC CSL-binding site. The vertical dividing line separates lanes run noncontiguously on the same gel. (C) Quantitative data of ChIP assays in B. Results are shown as fold enrichment of anti-NOTCH1–immunoprecipitated DNA over DNA immunoprecipitated with control Abs. Bars represent mean values ± SEM (n = 3). Ctrl, control. (D) Luciferase reporter assay of SupT1 T-ALL cells cotransfected with a reporter vector containing the indicated combinations of WT and mutated (mut) sequences corresponding to the 5 putative CSL-binding sites identified in CD44, along with either retrovirus encoding ICN1 together or not with dnMAML1-encoding retrovirus or with GFP-only retrovirus as control. Data are represented as mean fold induction SEM over control cells as in A (n = 3). Data shown in A and D were corrected by FDR. (E) Chromatin landscapes upstream of the CD44 locus were derived from ChIP-seq raw data of human CUTLL1 cells under steady-state (basal), Notch-off, and Notch-on conditions (GEO GSE51800), previously described by Wang et al. (23). The presence of a 5′ distal enhancer containing a dynamic NOTCH1-PBPJ site and a RUNX1 site is shown. H3K27ac peaks associated with this dynamic NOTCH1 site are shown. *P < 0.05; **P < 0.01; ***P < 0.001.