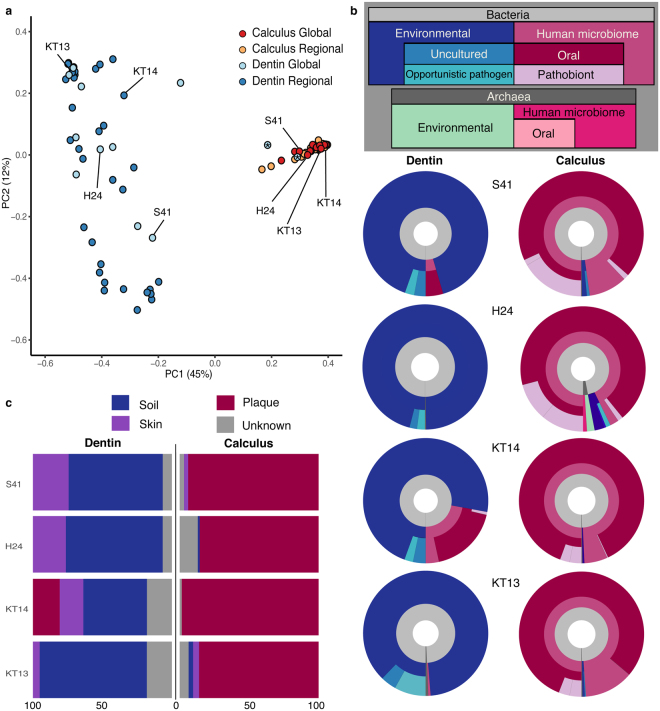
Figure 3.
Microbial communities represented in archaeological dental calculus and dentin are distinct. (a) Principal Coordinates Analysis (PCoA) of Bray-Curtis distances of all bacterial and archaeal species-level assignments from dental calculus and dentin. Color indicates material type (dental calculus or dentin) and membership in global or regional dataset. Microbial taxonomy was assigned using MALT with the full NCBI nucleotide database. Dentin samples marked with an asterisk belong to individuals NF47 and NF217 in the global dataset, and may represent teeth with carious lesions (Supplementary Figure S2). (b) A subset of four sample pairs from the global (S41, H24) and regional (KT14, KT13) sample sets were selected to further demonstrate differences in dental calculus and dentin microbial communities. Species represented in the MALT results were sorted into a layered classification scheme (see legend in gray box), and the proportions of reads assigned to each taxon were used to generate Krona plots. (c) Stacked bar plots of Bayesian SourceTracker results for the four selected pairs display estimated proportions of source contribution at the genus level, using modern plaque, skin, and soil datasets as model sources. Both approaches show overwhelming abundance of environmental bacteria within dentin samples, while most microbial DNA within the paired calculus samples are associated with, and most likely derive from, the human oral microbiome. The robustness of the human microbiome signal to the environmental signal is driving the separation we see in the PCoA.