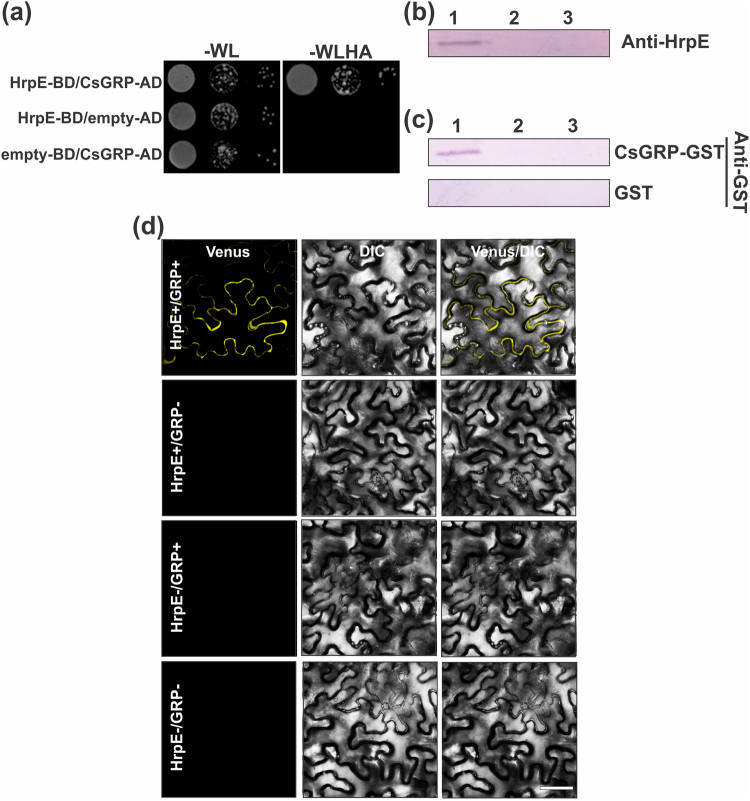
Figure 4.
Yeast Two Hybrid, Far Western Blot and BiFC assays of the interaction between HrpE and CsGRP. (a) Representative photograph of YTH assay showing that HrpE interacts with CsGRP. Yeast cells were co-transformed with: HrpE-BD/CsGRP-AD, HrpE-BD/empty-AD and empty-BD/CsGRP-AD. Yeast growth (serial 1:10 dilutions) is shown in −WL plates and in −WLHA plates containing 35 mM 3AT. (b) Purification of the Hrp-pilus from XcchrpG+ (lane 1) and XcchrpG− (lane 2) grown in XVM2 medium and from XcchrpG+ (lane 3) grown in SB. Proteins obtained from pilus preparations were analyzed by Tricine-SDS-PAGE and Western blot revealed with anti-HrpE polyclonal antibody. (c) Far Western blots showing interactions between HrpE present in the Hrp-pilus preparations and CsGRP-GST. Nitrocellulose membranes similar to that showed in (b) were incubated with 50 μg CsGRP-GST or GST as a control and, after washing, probed with anti-GST polyclonal antibody. (d) Confocal laser-scanning micrographs of the abaxial surface of N. benthamiana leaves. BiFC constructs of HrpE-nVenus/CsGRP-cCFP (HrpE+/GRP+), HrpE-nVenus/empty-cCFP (HrpE+/GRP−), empty-nVenus/GRP-cCFP (HrpE−/GRP+), and empty-nVenus/empty-cCFP (HrpE−/GRP−) were co-expressed in N. benthamiana using agroinfiltration. Scale bars represent 25 μm. DIC: Differential Interference Contrast.