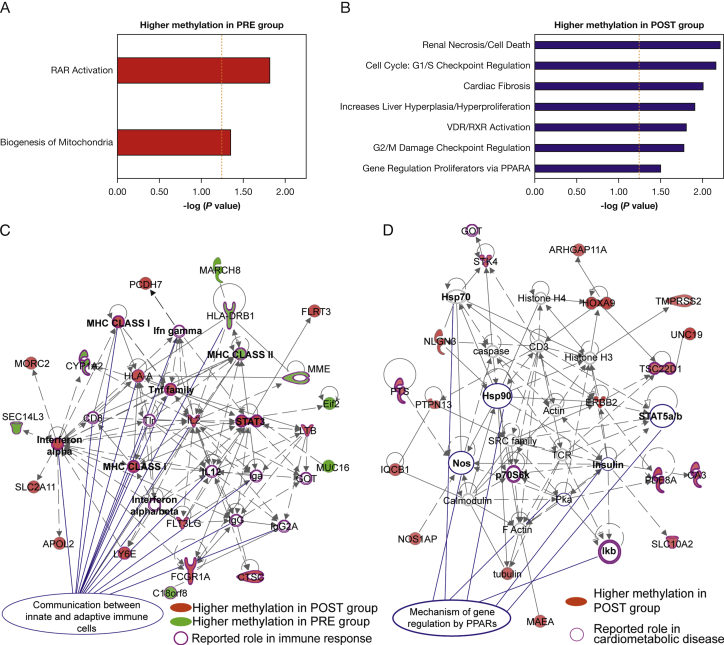
Figure 3.
Pathway and biological processes associated with DMRs. Biologically relevant gene interaction networks were identified by statistically significant overrepresentation in genes associated with the DMRs. Pathways and biological processes associated with DMRs show higher DNA methylation in the PRE group (red bars, panel A) and POST group (blue bars, panel B). Vertical orange dashed bars depicts the significance cutoff value for the overrepresentation test [–log10(P = .05) = 1.3; hypergeometric test). C, DMR-associated gene networks. Genes associated with DMRs with higher DNA methylation in POST and PRE samples are shown in red and green, respectively. Molecules with a reported function in immune response are circled in purple. Molecules with a role in communication between immune cells are indicated with blue lines. D, Gene network corresponding to mechanisms of gene regulation by peroxisome proliferation-activated receptors (PPARs) overrepresented in DMRs with high DNA methylation in the POST group (shown in red). Molecules with a reported role in cardiometabolic diseases are circled in purple. Molecules associated with PPAR pathways are indicated with blue lines. PPARA = peroxisome proliferation-activated receptor α; RAR = retinoic acid receptor; RefSeq = Reference Sequence; VDR/RXR = vitamin D receptor/retinoid X receptor. See Figure 1 legend for expansion of other abbreviations.