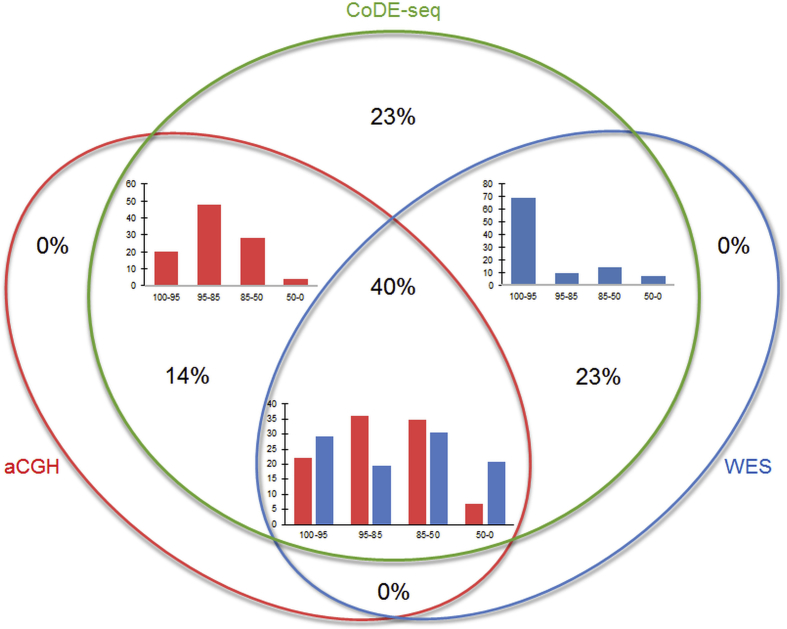
Figure 2.
CNV detection through CoDE-seq, aCGH or whole-exome sequencing (WES) in the 40 participants for whom aCGH data were available. CoDE-seq detected 181 CNVs; aCGH detected 97 CNVs and whole-exome sequencing detected 114 CNVs. Histograms show the frequency (%) of the CNVs detected by aCGH (red histograms) or by whole-exome sequencing (blue histograms) according to their size (100–95: between 95 and 100% of the size of the CNVs detected by CoDE-seq; 95–85: between 85 and 95% of the size of the CNVs detected by CoDE-seq; 85–50: between 50 and 85% of the size of the CNVs detected by CoDE-seq; 50–0: below 50% of the size of the CNVs detected by CoDE-seq). CNV frequency (%) in each part of the Venn diagram is also shown.