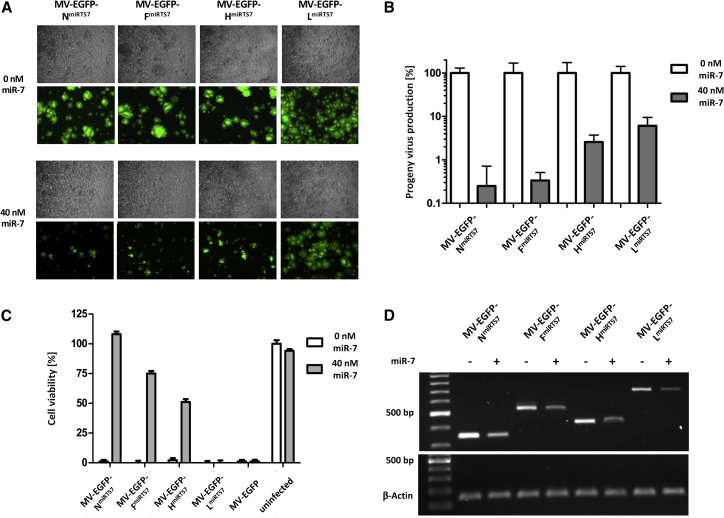
Figure 3.
Functional Analysis and Comparison of Different miRTS Box Positions
Vero cells were transfected with 0 or 40 nM miR-7-5p mimics and subsequently infected with MV-EGFP-NmiRTS7/-FmiRTS7/-HmiRTS7/-LmiRTS7 or MV-EGFP at an MOI of 0.03. (A) Fluorescence and phase contrast microscopy images 34 hr post-infection, ×50 magnification. (B) Progeny virus determination. Thirty-six hours post-infection, cells were scraped into their medium. Virus progeny titers were determined and are shown with normalization to progeny virus titers in absence of transfected microRNA. Error bars represent SD of three technical replicates per sample. (C) Cell viability of all samples was determined at 96 hr post-infection using a colorimetric (XTT) assay. Error bars represent SD of three technical replicates per sample. (D) Total RNA was isolated 32 hr post infection, subjected to RT-PCR, and cDNA was used for PCR. Gene-specific primer pairs corresponding to regions up- and downstream of the miRTS box insertion sites within the N, F, H, and L ORF, respectively, were used (upper gel), while β-actin-specific primers were used as an input control (lower gel). RT-PCR products were subjected to agarose gel electrophoresis.