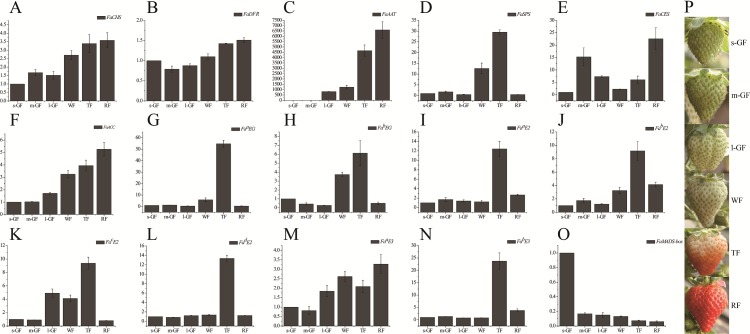
Figure 5. Expression profile of candidate genes during different fruit development and ripening stages in qRT-PCR.
qRT-PCR analysis of strawberry candidate gene (A) FaCHS; (B) FaDFR; (C) FaAAT; (D) FaSPS; (E) FaCES; (F) FaACC; (G) FaaEG; (H) FabEG; (I) FaaE2; (J) FabE2; (K) Fa cE2; (L) Fa dE2; (M) Fa aE3; (N) Fa bE3; (O) FaMADS-box. (P) Tissues of strawberry ‘Toyonoka’ used in qRT-PCR. CHS, chalcone synthase; DFR, bifunctional dihydroflavonol 4-reductase; AAT, alcohol acyltransferase; SPS, sucrose-phosphate synthase 1; CES, cellulose synthase A catalytic sub-unit 4; ACC, Acetyl-coenzyme A carboxylase carboxyl transferase sub-unit alpha; aEG, endoglucanase CX-like; bEG, endoglucanase 24-like; aE2, ubiquitin-conjugating enzyme E2 5-like; bE2, ubiquitin-conjugating enzyme E2 23-like; cE2, ubiquitin-conjugating enzyme E2 28-like; dE2, ubiquitin-conjugating enzyme E2 4-like; aE3, E3 ubiquitin-protein ligase UPL3-like; bE3, cullin-1-like; MADS-box, MADS-box protein ZMM17-like. FaActin were used as an internal control. Result shows expression value of candidate genes relative to s-GF stage. The experiments were repeated three times and provided consistent results. The mean values and error bars were obtained from three biological and three technical replicates.