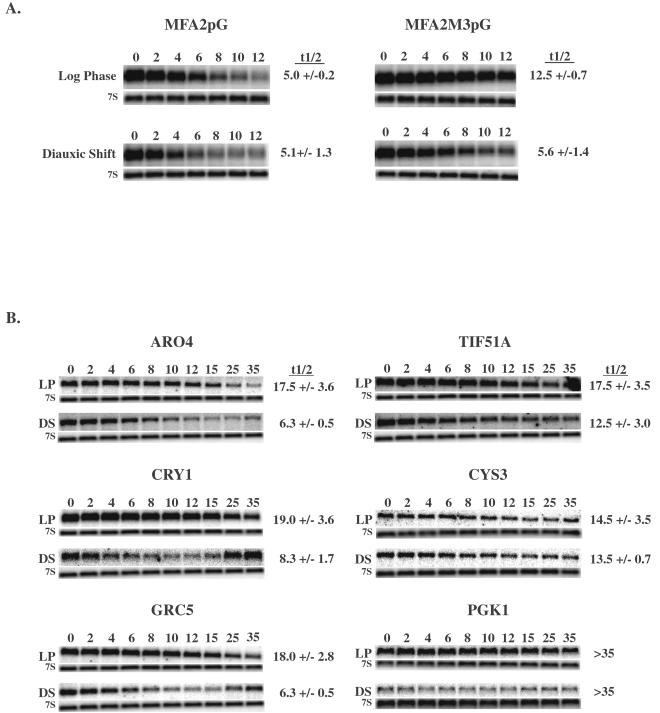
Figure 1.
Multiple mRNAs are destabilized at diauxic shift. (A) mRNA half-life analysis of MFA2pG and MFA2M3pG mRNA from either log-phase or diauxic shift cultures. 5′ end-labeled oRP127 was used to detect mRNAs (upper panel). The numbers above the lanes indicate the number of minutes after transcription repression. As a loading control, blots were stripped and reprobed with oRP100 to detect the RNA polymerase III 7S transcript (lower panel). (B) mRNA half-life measurements of six different mRNAs at log phase (LP) or diauxic shift (DS). The numbers above the lanes indicate the number of minutes after transcription repression. The ARO4, CRY1, GRC5, CYS3, and TIF51A mRNAs were detected with the use of 5′ end-labeled oligonucleotides complementary to their coding regions (see MATERIALS AND METHODS). The PGK1 mRNA was detected with a random prime-labeled probe that is complementary to the 5′ UTR and 171 nucleotides of the coding region. The oligonucleotide used to detect CRY1 mRNA is also complementary to CRY2 mRNA, which is much lower in abundance (Paulovich et al., 1993). The oligonucleotide used to detect TIF51A (HYP2) is also complementary to the TIF51B (ANB1, HYP1) mRNA, which is not expressed under these aerobic conditions (Schnier et al., 1991). 7S RNA again was used as a loading control (7S). In both A and B, mRNA half-life values were calculated by normalization to the 7S RNA and are the mean and SD of at least two experiments.