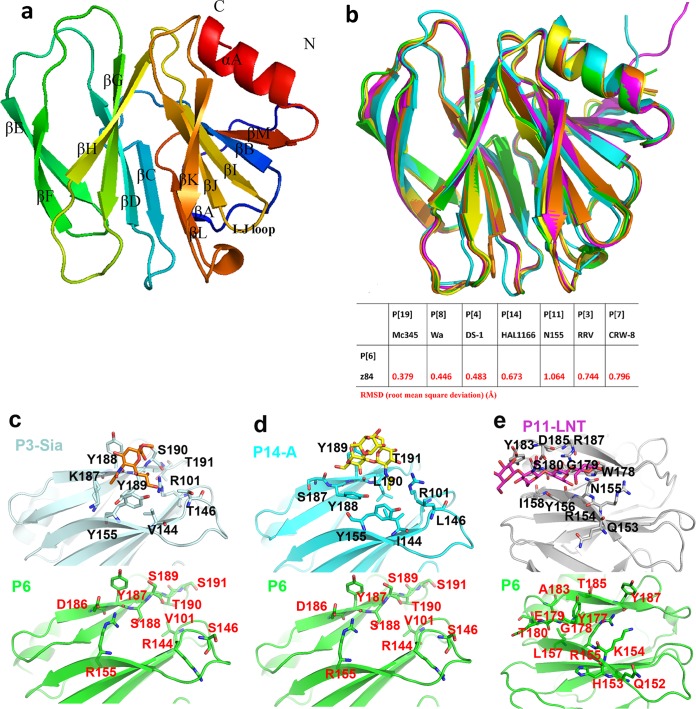
FIG 3.
Structural characteristics of P[6] VP8*. (a) Overall structure of porcine P[6] VP8* with two twisted antiparallel sheets consisting of strands A, L, D, G, and H and M, B, I, J, and K. (b) (Top) Superimposition of porcine P[6] VP8* structure (PDB ID 5YMU; green) on those of P[19] (PDB ID 5GJ6; pale cyan), Wa P[8] (PDB ID 2DWR; yellow), DS-1 P[4] (PDB ID 2AEN; orange), and HAL1166 P[14] (PDB ID 4DRV; cyan). (Bottom) Structure comparison of porcine P[6] (5YMU), RRV P[3] (1KQR), CRW-7 P[7] (2I2S), HAL1166 P[14] (4DRV), DS-1 P[4] (2AEN), Wa P[8] (2DWR), HRV P[11] (4YG0), andMc345 P[19] (5GJ6). The values in the table represent RMSDs (in Å) of the Cα atoms of one VP8* monomer. VP8* structures are shown as a cartoon. (c) Comparison of the residues involved in sialic acid binding in P[3] VP8* (PDB ID 1KQR) (top; pale cyan) and that of porcine P[6] VP8* structure (PDB ID 5YMU) (bottom; green). The amino acids involved in the interaction are shown in stick representation. (d) Comparison of the residues involved in A-type HBGA binding in P[14] VP8* (PDB ID 4DRV) (top; cyan) and that of porcine P[6] VP8* structure (PDB ID 5YMU) (bottom; green). (e) Comparison of the residues involved in LNT binding in P[11] VP8* (PDB ID 4YFZ) (top; gray) and that of porcine P[6] VP8* structure (PDB ID 5YMU) (bottom, green).