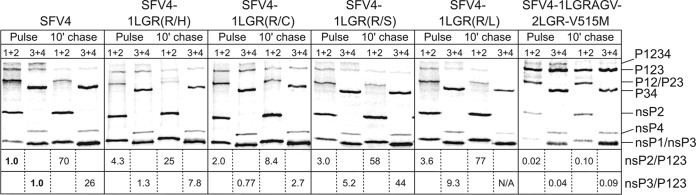
FIG 2.
Processing of the ns-polyproteins in BHK-21 cells infected with SFV4 and its mutant variants. Cells were infected with wt SFV4 and five independently plaque-purified isolates of mutants and labeled with [35S]methionine, followed by a 10-min chase with an excess of cold methionine. The proteins were extracted from the infected cells, denatured with 1% SDS, analyzed via immunoprecipitation with antibodies against nsP1 and nsP2 or against nsP3 and nsP4 (shown above the lines) followed by SDS-PAGE, and visualized using a Typhoon imager; quantification was performed using ImageQuant software. The viruses used in the assays are indicated at the top of each panel, and the origin of the samples (pulse or chases) is shown below. The positions of the SFV nsPs and their polyprotein precursors are indicated on the right. The ratios indicate the amount of label that was incorporated into individual nsP2s or nsP3s compared with the amount incorporated into unprocessed P123; ratios for SFV4 in pulsed samples are taken as 1.