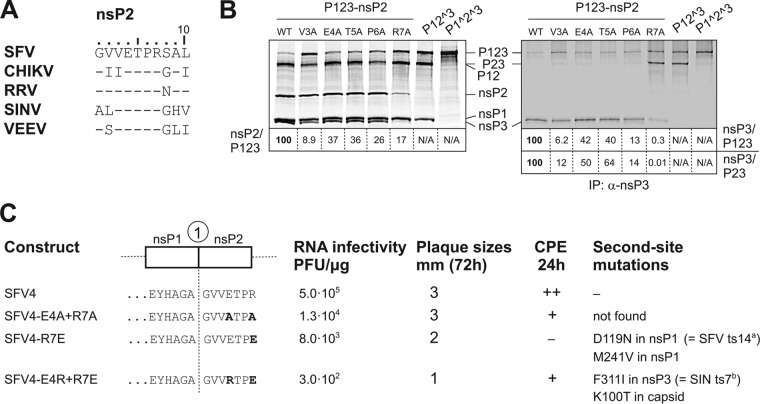
FIG 5.
The N-terminal amino acid residues of SFV nsP2 affect polyprotein processing, RNA infectivity, virus growth, and cytotoxicity of the corresponding mutant viruses. (A) Comparison of sequences of first 10 N-terminal amino acid residues of SFV, CHIKV, RRV, SINV, and VEEV nsP2. (B) Left panel, results of in vitro transcription/translation/processing of P123 (wt), P123-V3A, P123-E4A, P123-T5A, P123-P6A, P123-R7A, P12^3, and P1^2^3 of SFV. Right panel, analysis of the same samples after immunoprecipitation with anti-nsP3 antibodies. Proteins were separated by SDS-PAGE and visualized using a Typhoon imager; quantification was performed using ImageQuant software. The ratios under the left panel indicate the amount of label that was incorporated into the individual nsP2 protein compared with the amount incorporated into unprocessed P123. The ratios under the right panel indicate the amount of label that was incorporated into the individual nsP3 protein compared with the amount incorporated into unprocessed P123 or unprocessed P23. In all cases, ratios for wt P123 are taken as 100. (C) Graphical presentation of the 1/2 site region of SFV4 and its mutant versions. Mutated residues of nsP2 are shown in boldface. The results for RNA infectivity, plaque sizes, cytopathic effects (CPE) at 24 hpi, and second-site mutations revealed in the genomes of the rescued mutant viruses are shown. All experiments were repeated at least twice with similar results.