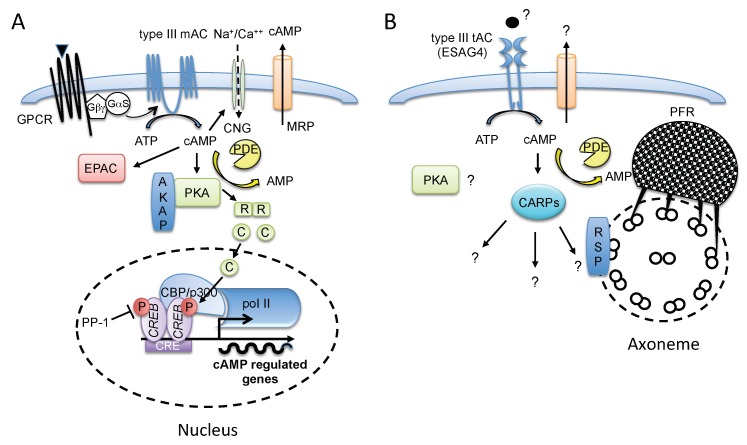
Figure 1.
Schematic representation of cAMP signaling in mammalian cells (A) vs. T. brucei (B), highlighting the contrast between canonical mammalian cAMP/PKA signaling pathway and the African trypanosomes’ cAMP signaling, which mainly concentrates in the flagellum and is characterized by the almost total absence of polII transcriptional regulation. (A) In mammals, ligand (triangle) binding activates GPCR, which undergoes a conformational change and then activates the G proteins by promoting the exchange of GDP/GTP associated with the Gα subunit, triggering its dissociation from the Gβ/Gγ dimer to activate type III AC. AC produces cAMP from ATP. High local levels of cytosolic cAMP lead to activation of PKA holoenzyme, which binds the AKAP through a hydrophobic dimerization domain of the PKA-R subunit, Epac or CNG channel. Upon cAMP binding to PKA-R, PKA-C subunits dissociate, then translocate to the cell nucleus, and induce the phosphorylation of transcription factors, such as CREB, to activate cAMP-driven genes. CREB inactivation is promoted by a phosphatase (e.g., PP-1). PDE and MRP decrease intracellular cAMP levels and counterbalance the intracellular cAMP effect. (B) In T. brucei, a putative ligand (circle) or membrane stress (hypotonic, acidic, proteolytic) activates flagellar type III AC (prototype ESAG4 in bloodstream form). This AC is topologically similar to receptor-type GC and produces cAMP from ATP. In T. brucei no classical PKA effector is activated by cAMP; instead, the cAMP targets are CARPs, components of unknown function, which participate in a putative novel cAMP signaling pathway. RSP represents an AKAP-like protein linked to the flagellar axoneme (RSP3/AKAP97-like); PFR represents the parafagellar rod structure of the flagellum, which is linked to PDEs (TbPDEB1/B2). No CNG channels or Epac have been characterized in trypanosomatids, and there is no evidence for cAMP secretion via membrane channels. AKAP, A-kinase anchoring protein; CBP, cAMP-binding protein; CARP, cAMP response protein; CNG, cyclic nucleotide-gated ion channel; CRE, cAMP-response elements; CREB, cAMP response element-binding protein; EPAC, exchange protein directly activated by cAMP; Gαs, stimulatory G protein alpha subunit; Gβγ, G protein beta gamma subunits; GPCR, G-protein-coupled receptor; MRP, multidrug resistance protein; PDE, phosphodiesterase; PFR, paraflagellar rod; PKA, protein kinase A; PolII, RNA polymerase II; RSP, radial spoke protein.