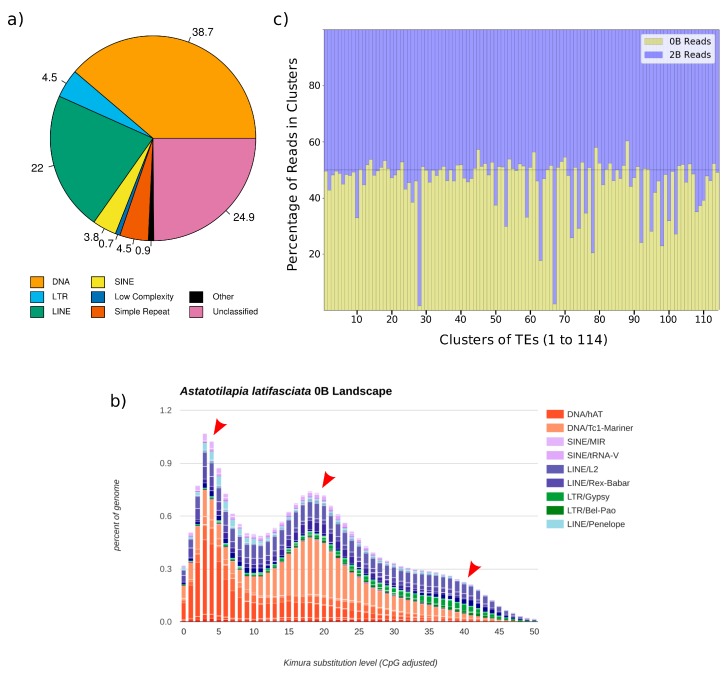
Figure 1.
Genomic characteristics of repeated DNAs in the Astatotilapia latifasciata genome. (a) Percentage of each type of repetitive sequence in the total repetitive portion detected by RepeatMasker on the B- genome. Numbers in this figure represent the percentage of each repeat group compared to the total repetitive content, rather than their percentage in the genome. DNA transposons and long interspersed nuclear element (LINEs) dominate the repetitive content. Unclassified sequences comprise mostly multigenic families and gene clusters. (b) Repeat landscape of the A. latifasciata genome. For each element, the graph shows the sequence divergence from its consensus (x-axis) in relation to the number of copies on the genome (y-axis). Peaks represent insertion waves (red arrows) of elements into the genome. Elements with older insertion waves are shown on the right side of the graph, while newer insertions are depicted on the left side. Different colors show distinct element types, as described at the right. For a detailed and interactive version of the graph, please refer to Supplemental file 2. (c) Comparative analysis of repeats on the B- (0B) and B+ (2B) genomes via RepeatExplorer. Each column represents 100% of the reads in a cluster; the read proportions from the B- (0B) and B+ (2B) genomes are shown in yellow and blue, respectively. Clusters with higher proportions of B+ (2B) reads (in blue) are shown by an expansion of the blue color into the lower part of the graph (yellow). LINE: Long interspersed nuclear element; LTR: Long terminal repeat; SINE: Short interspersed nuclear element; TE: Transposable element.