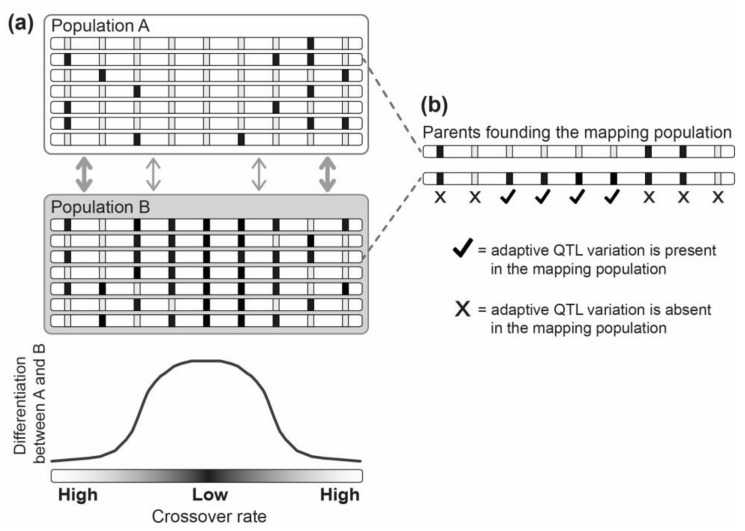
Figure 4.
Although called ‘quantitative trait locus’ (QTL) mapping, this method cannot distinguish between locus and allele-only clustering. (a) Shown are two exemplary populations adapted to different local environments under genetic exchange. Each population is represented by seven individuals, and each individual is illustrated as a single haploid chromosome. The boxes within the chromosomes indicate positions of loci influencing locally adaptive trait variation (relevant loci), while the different gray tones of the boxes represent the alternative local alleles. We assume that all relevant loci have the same selective effect size and that they have not evolved to cluster within the genome in response to gene flow (such a priori knowledge on the distribution of relevant loci is, of course, absent in a real study). In the illustrated case, local adaptation with gene flow is therefore facilitated only by fortuitous co-localization of relevant loci within the low-crossover chromosome center (the thickness of the gray arrows indicates the degree of effective gene flow between the populations). The result is stronger differentiation in the chromosome center because of allele-only clustering. (b) When the parental individuals used for a QTL cross are sampled from the two depicted populations, genetic variation at relevant QTL, and therefore QTL detection in general, will be biased towards the high-differentiation chromosome region. Overall, this highlights that interpretations of QTL mapping results should consider the degree to which the parental mapping populations are differentiated and connected by gene flow (see also [107]).