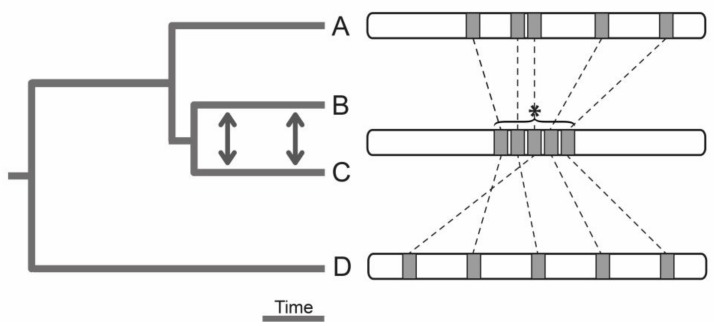
Figure 5.
Conceptual outline for the inference of a genome structure evolved in response to gene flow. Compared are de novo genome assemblies from multiple populations or related species (hereafter simply ‘species’) evolved under various degrees of genetic exchange. If a genome region important for local adaptation with gene flow is known a priori (here identified between species B and C, and denoted by an asterisk), the degree of genomic clustering of homologous sequences in a related species that evolved without gene flow (species A) should be reduced to be indicative of an adaptive response of the genome structure to gene flow (see also [112]). Such a test should account for the degree of ‘neutral’ overall structural change of a genome and should profit from integrating distantly related species (species D) because the origin of adaptive structural genome variation can be old and pre-date recent species splits [71]. Without a priori knowledge of genome regions important for local adaptation with gene flow, an evolved genome structure in response to gene flow may be detectable by more abundant structural genome differences between species evolved with versus without gene flow. Finally, if genome regions with lower crossover rates are indeed co-localization hotspots of relevant loci, the ratio of total structural change in low to high crossover genome regions should be higher between species evolved with versus without gene flow.