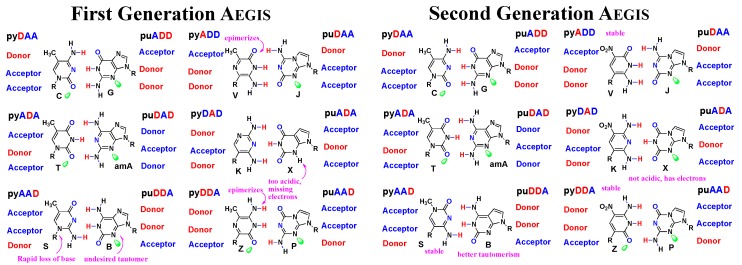
Figure 1.
Examples of artificially expanded genetic information system (AEGIS) xNA building blocks in their “first generation” forms (left) and their “second generation” forms (right). AEGIS exploits the two rules that guide standard Watson–Crick nucleobase pairing: (a) size (large purines with small pyrimidines) and (b) hydrogen bonding (hydrogen bond acceptors A, with hydrogen bond donors D). Rearranging A and D groups gives an artificially expanded genetic information system (AEGIS) with up to 12 nucleotides forming six orthogonal pairs, with a functionality not present in standard DNA. AEGIS Z, for example, carries a nitro group (enhance the intrinsic binding potential of AEGIS libraries, and allowing the nucleobase to act as a general acid-base).