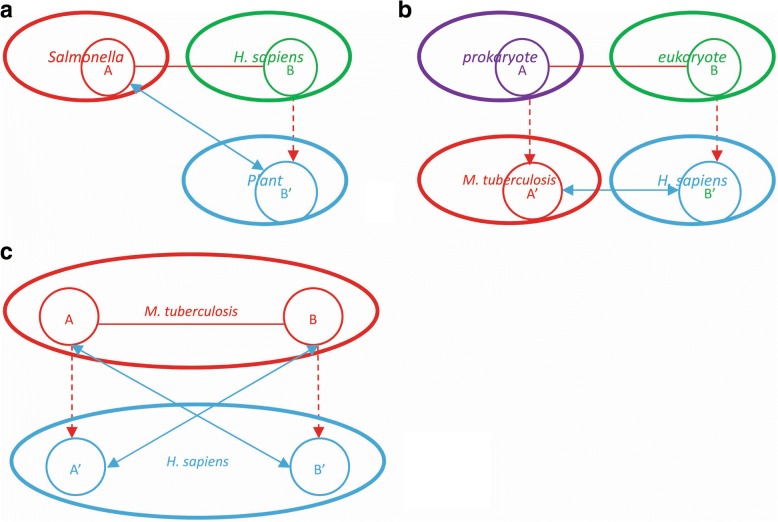
Fig. 7.
Illustration of the way of deriving interlogs. The ellipses in red and blue denote the target species and the ellipses in other colors denote the source species. The circles denote genes. The red full line denotes the experimental protein-protein interactions. The red dotted line denotes ortholog mapping. The blue full line with arrows at two ends denotes the derived interlogs. The methods illustrated in (a) and (b) exploited the pathogen-host PPIs of other species to derive the interlogs of the target species, while the method illustrated in (c) transferred the knowledge of intra-species M. tuberculosis H37Rv PPI networks to predict protein interactions between M. tuberculosis H37Rv and H. sapiens