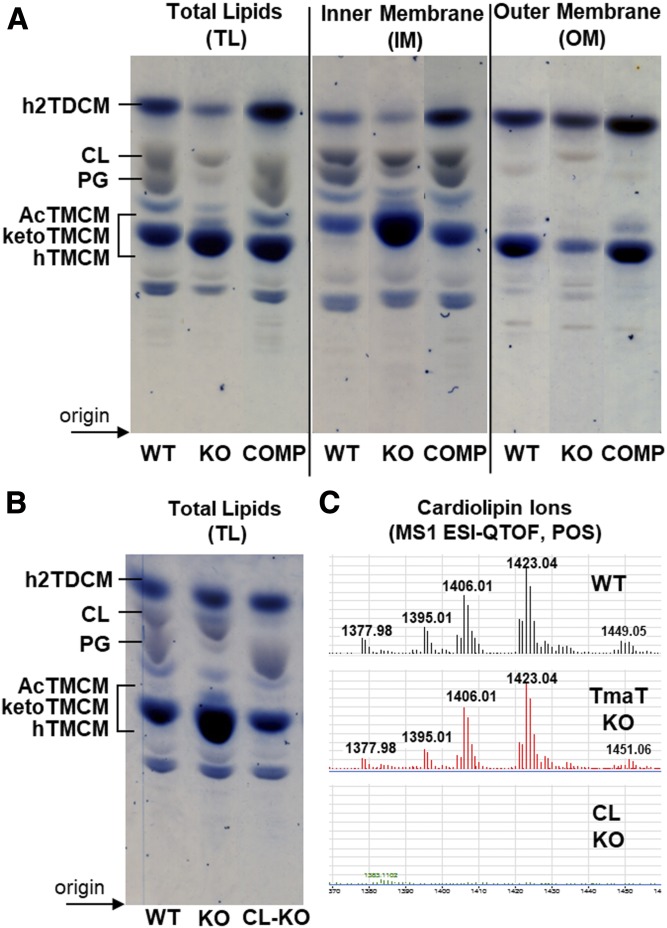
Fig. 6.
HPTLC analysis of lipids from C. glutamicum WT and the Δtmat mutant. A: TL extracts, as well as lipids derived from IM and OM fractions from C. glutamicum WT, the Δtmat mutant (KO), and the complementation strain (COMP) were analyzed by HPTLC. Glycolipids (blue) and phospholipid (gray) were detected by orcinol-sulfuric acid staining. Select TLC lanes are shown from replicate analyses (supplemental Fig. S1). B: A C. glutamicum mutant lacking CL synthase (NCgl2646) was generated, and TLs were analyzed by HPTLC. C: LC-MS of lipids from C. glutamicum WT bacteria, the ΔtmaT, and the ΔNCgl2646 mutants. Only the region of the LC chromatogram showing CL species is shown.