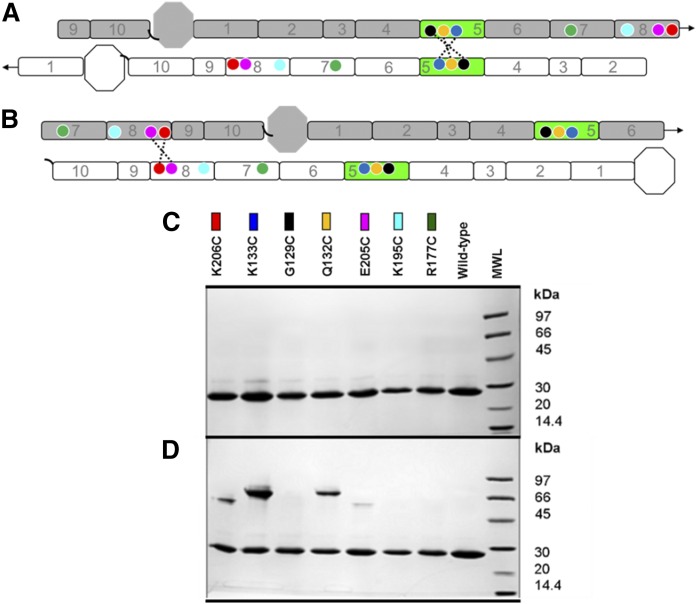
Fig. 2.
Design of mutants for the postulated APOA1 registries in discoidal rHDL. A: A 2D representation of the molecular interactions between two antiparallel APOA1 monomers (imagine two APOA1 belts pulled off the edge of a particle and laid flat). Helix 5 (H5, green) of APOA1 (gray) is shown opposite to H5 of its antiparallel APOA1 partner molecule (white). Circles represent the positions of APOA1 residues K133 (dark blue), G129 (black), Q132 (yellow), K206 (red), E205 (pink), K195 (cyan), and R177 (green). The dotted line shows disulfide bonds predicted to form with APOA1 mutant K133C, G129C, and Q132C in the 5/5 registry. Arrows at the end of the molecules show the continuity of the APOA1 disc helices [N terminus (octagon), H1 through H10, C terminus (black, curved line)]. B: A 2D representation of two antiparallel APOA1 monomers with H5 (green) of APOA1 (gray) opposite to H2 of its APOA1 partner molecule (white). Circles represent the same residues indicated above. The dotted line shows the disulfide bond predicted to form in APOA1 mutant K206C and E205C in a 5/2 registry. C: SDS-PAGGE analysis of WT APOA1 and Cys-mutants reconstituted into rHDL particles under reducing conditions (3 mM DTT). Colored rectangles correspond to the positions of the residues on APOA1 diagrams in A and B. D: SDS-PAGGE (nonreducing) analysis of WT APOA1 and Cys-mutant rHDL after incubation at 21°C for 48 h in the absence of a reducing agent.