Figure 7. Induced dimerization of CEACAM3 has no effect on binding to HopQ.
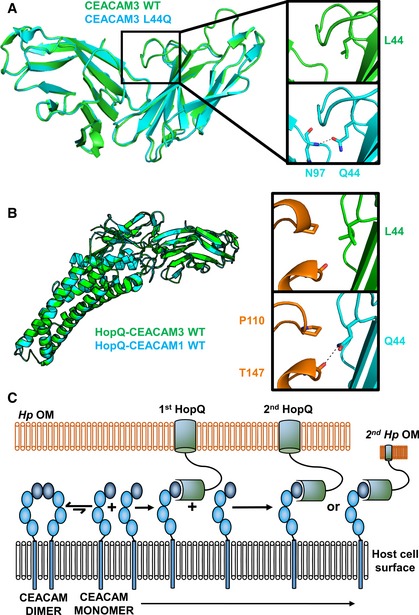
- CEACAM3 WT (green) at position 44 is a leucine. By AUC, it is monomeric; however, it adopts a dimeric conformation in the crystal structure. CEACAM3 Leu44Gln mutation (cyan) creates two symmetrical hydrogen bonds across the dimer interface to the backbone of Asn97. This causes dimerization of CEACAM3. No structural changes are observed between CEACAM3 and mutant with a RMSD of 0.33 Å.
- Superposition of the HopQ‐CEACAM1 complex (cyan) with the HopQ‐CEACAM3 WT complex reveals little structural changes between the complexes with a RMSD of 0.30 Å. In WT CEACAM3 (green), Leu44 forms van der Waal interactions with Pro110 of HopQ (orange). In CEACAM3, Leu44Gln (cyan) forms van der Waal interactions with Pro110 and a hydrogen bond to the main chain of Thr147 (orange).
- Model of HopQ recruitment of CEACAM. CEACAM1 (blue) is predominately dimeric. HopQ (green) binding of CEACAM monomers causes redistribution of dimers to monomers which can bind further HopQ on the same bacterium or recruit other bacteria to the host cell.
